Electrochemiluminescent Arrays For Toxicity Screening
- PMID: 24639601
- PMCID: PMC3955387
Electrochemiluminescent Arrays For Toxicity Screening
Figures







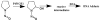
References
-
- Březina M, Zuman P. Polarography in Medicine, Biochemistry, and Pharmacy. Interscience; New York: 1958.
-
- Ramsay G, editor. Commercial Biosensors. Wiley; New York: 1998.
-
- Debad JB, Glezer EN, Leland JK, Sigal GB, Wholstadter J. In: Electrogenerated Chemiluminescence. Bard AJ, editor. Marcel Dekker; NY: 2004. pp. 359–396.
-
- Miao W, Bard AJ. Anal Chem. 2004;76:5379. - PubMed
- Wang X, Bobbitt DR. Anal Chim Acta. 1999;383:213.
- Hendrichkson HP, Anderson P, Wang X, Pittma Z, Bobbitt DR. Microchem J. 2000;65:189.
-
- Miao W, Choi JP, Bard AJ. J Am Chem Soc. 2002;124:14478. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
