Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species
- PMID: 24640988
- PMCID: PMC4006885
- DOI: 10.1021/ac500447w
Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species
Abstract
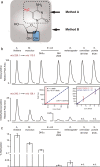
The methylation of cytosine to 5-methylcytosine (5-meC) is an important epigenetic DNA modification in many bacteria, plants, and mammals, but its relevance for important model organisms, including Caenorhabditis elegans and Drosophila melanogaster, is still equivocal. By reporting the presence of 5-meC in a broad variety of wild, laboratory, and industrial yeasts, a recent study also challenged the dogma about the absence of DNA methylation in yeast species. We would like to bring to attention that the protocol used for gas chromatography/mass spectrometry involved hydrolysis of the DNA preparations. As this process separates cytosine and 5-meC from the sugar phosphate backbone, this method is unable to distinguish DNA- from RNA-derived 5-meC. We employed an alternative LC-MS/MS protocol where by targeting 5-methyldeoxycytidine moieties after enzymatic digestion, only 5-meC specifically derived from DNA is quantified. This technique unambiguously identified cytosine DNA methylation in Arabidopsis thaliana (14.0% of cytosines methylated), Mus musculus (7.6%), and Escherichia coli (2.3%). Despite achieving a detection limit at 250 attomoles (corresponding to <0.00002 methylated cytosines per nonmethylated cytosine), we could not confirm any cytosine DNA methylation in laboratory and industrial strains of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Saccharomyces boulardii, Saccharomyces paradoxus, or Pichia pastoris. The protocol however unequivocally confirmed DNA methylation in adult Drosophila melanogaster at a value (0.034%) that is up to 2 orders of magnitude below the detection limit of bisulphite sequencing. Thus, 5-meC is a rare DNA modification in drosophila but absent in yeast.
Figures
References
-
- Tamaru H.; Selker E. U. Nature 2001, 414, 277–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

