Metabolic changes associated with the long winter fast dominate the liver proteome in 13-lined ground squirrels
- PMID: 24642758
- PMCID: PMC4042184
- DOI: 10.1152/physiolgenomics.00190.2013
Metabolic changes associated with the long winter fast dominate the liver proteome in 13-lined ground squirrels
Abstract
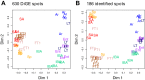
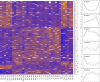
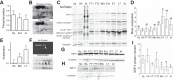
Small-bodied hibernators partition the year between active homeothermy and hibernating heterothermy accompanied by fasting. To define molecular events underlying hibernation that are both dependent and independent of fasting, we analyzed the liver proteome among two active and four hibernation states in 13-lined ground squirrels. We also examined fall animals transitioning between fed homeothermy and fasting heterothermy. Significantly enriched pathways differing between activity and hibernation were biased toward metabolic enzymes, concordant with the fuel shifts accompanying fasting physiology. Although metabolic reprogramming to support fasting dominated these data, arousing (rewarming) animals had the most distinct proteome among the hibernation states. Instead of a dominant metabolic enzyme signature, torpor-arousal cycles featured differences in plasma proteins and intracellular membrane traffic and its regulation. Phosphorylated NSFL1C, a membrane regulator, exhibited this torpor-arousal cycle pattern; its role in autophagosome formation may promote utilization of local substrates upon metabolic reactivation in arousal. Fall animals transitioning to hibernation lagged in their proteomic adjustment, indicating that the liver is more responsive than preparatory to the metabolic reprogramming of hibernation. Specifically, torpor use had little impact on the fall liver proteome, consistent with a dominant role of nutritional status. In contrast to our prediction of reprogramming the transition between activity and hibernation by gene expression and then within-hibernation transitions by posttranslational modification (PTM), we found extremely limited evidence of reversible PTMs within torpor-arousal cycles. Rather, acetylation contributed to seasonal differences, being highest in winter (specifically in torpor), consistent with fasting physiology and decreased abundance of the mitochondrial deacetylase, SIRT3.
Keywords: Ictidomys tridecemlineatus; autophagy; mitochondria; starvation.
Copyright © 2014 the American Physiological Society.
Figures




References
-
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284, 2001. - PubMed
-
- Breiman L. Random forests. Machine Learn 45: 5–32, 2001.
-
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev 83: 1153–1181, 2003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

