A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots
- PMID: 24642943
- PMCID: PMC4001404
- DOI: 10.1105/tpc.113.122069
A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots
Abstract
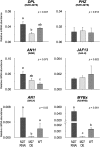
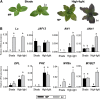
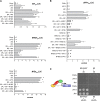
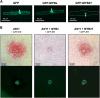
Plants require sophisticated regulatory mechanisms to ensure the degree of anthocyanin pigmentation is appropriate to myriad developmental and environmental signals. Central to this process are the activity of MYB-bHLH-WD repeat (MBW) complexes that regulate the transcription of anthocyanin genes. In this study, the gene regulatory network that regulates anthocyanin synthesis in petunia (Petunia hybrida) has been characterized. Genetic and molecular evidence show that the R2R3-MYB, MYB27, is an anthocyanin repressor that functions as part of the MBW complex and represses transcription through its C-terminal EAR motif. MYB27 targets both the anthocyanin pathway genes and basic-helix-loop-helix (bHLH) ANTHOCYANIN1 (AN1), itself an essential component of the MBW activation complex for pigmentation. Other features of the regulatory network identified include inhibition of AN1 activity by the competitive R3-MYB repressor MYBx and the activation of AN1, MYB27, and MYBx by the MBW activation complex, providing for both reinforcement and feedback regulation. We also demonstrate the intercellular movement of the WDR protein (AN11) and R3-repressor (MYBx), which may facilitate anthocyanin pigment pattern formation. The fundamental features of this regulatory network in the Asterid model of petunia are similar to those in the Rosid model of Arabidopsis thaliana and are thus likely to be widespread in the Eudicots.
Figures




References
-
- Aharoni A., De Vos C.H.R., Wein M., Sun Z.K., Greco R., Kroon A., Mol J.N.M., O’Connell A.P. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28: 319–332. - PubMed
-
- Albert N.W., Arathoon S., Collette V.E., Schwinn K.E., Jameson P.E., Lewis D.H., Zhang H., Davies K.M. (2010). Activation of anthocyanin synthesis in Cymbidium orchids: variability between known regulators. Plant Cell Tissue Organ Cult. 100: 355–360.
-
- Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65: 771–784. - PubMed
-
- An X.H., Tian Y., Chen K.Q., Wang X.F., Hao Y.J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169: 710–717. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

