A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination
- PMID: 24642944
- PMCID: PMC4001371
- DOI: 10.1105/tpc.113.121830
A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination
Abstract
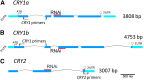
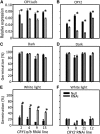
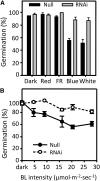
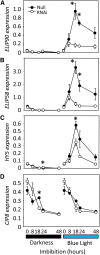
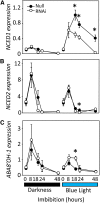
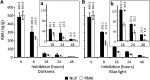
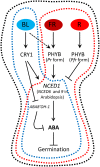
It is well known that abscisic acid (ABA) plays a central role in the regulation of seed dormancy and that transcriptional regulation of genes encoding ABA biosynthetic and degradation enzymes is responsible for determining ABA content. However, little is known about the upstream signaling pathways impinging on transcription to ultimately regulate ABA content or how environmental signals (e.g., light and cold) might direct such expression in grains. Our previous studies indicated that light is a key environmental signal inhibiting germination in dormant grains of barley (Hordeum vulgare), wheat (Triticum aestivum), and Brachypodium distachyon and that this effect attenuates as after-ripening progresses further. We found that the blue component of the light spectrum inhibits completion of germination in barley by inducing the expression of the ABA biosynthetic gene 9-cis-epoxycarotenoid dioxygenase and dampening expression of ABA 8'-hydroxylase, thus increasing ABA content in the grain. We have now created barley transgenic lines downregulating the genes encoding the blue light receptors CRYTOCHROME (CRY1) and CRY2. Our results demonstrate that CRY1 is the key receptor perceiving and transducing the blue light signal in dormant grains.
Figures

Comment in
-
Cryptochromes and seed dormancy: the molecular mechanism of blue light inhibition of grain germination.Plant Cell. 2014 Mar;26(3):846. doi: 10.1105/tpc.114.124727. Epub 2014 Mar 18. Plant Cell. 2014. PMID: 24642940 Free PMC article. No abstract available.
References
-
- Barrero J.M., Jacobsen J.V., Talbot M.J., White R.G., Swain S.M., Garvin D.F., Gubler F. (2012). Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 193: 376–386. - PubMed
-
- Bouly J.P., Schleicher E., Dionisio-Sese M., Vandenbussche F., Van Der Straeten D., Bakrim N., Meier S., Batschauer A., Galland P., Bittl R., Ahmad M. (2007). Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282: 9383–9391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

