Human papillomavirus genomics: past, present and future
- PMID: 24643174
- PMCID: PMC4430864
- DOI: 10.1159/000355952
Human papillomavirus genomics: past, present and future
Abstract
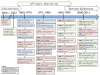
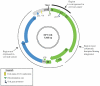
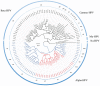
Human papillomaviruses (HPV) are a group of divergent DNA viruses, of which a select few evolutionarily related HPVs have emerged to be highly oncogenic and of significant medical importance. Essentially all cases of cervical cancer, as well as a subset of other anogenital and oral cancers are caused by this limited set of HPV types. At present, over 150 HPV types have been identified and may be classified into genera, species and types based upon comparison of the viral genome. Established nucleotide phylogenies sort the highly pathogenic HPV types to the genus Alphapapillomavirus (α-PV). A species group includes viral types with 60-70% genomic nucleotide similarity that share a most-recent common ancestor; for example the species group's alpha-9 (HPV16-related) and alpha-7 (HPV18-related), contain the majority of known oncogenic HPV types. Genomes from the same HPV type with 1-10% nucleotide differences designate HPV variant lineages. The established nucleotide variations observed in extant HPV genomes have been fixed through evolutionary processes prior to human population expansion and global dissemination. To characterize viral types and variants associated with pathology for clinical applications (e.g. screening), molecular epidemiological studies have proven essential for identifying links between HPV natural history and carcinogenicity. This chapter presents a historical account of HPV genomics in the context of major discoveries and advances over the past 2 thousand years.
© 2014 S. Karger AG, Basel.
Figures

References
-
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. - PubMed
-
- Herbst LH, Lenz J, Van Doorslaer K, et al. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology. 2009;383:131–5. - PubMed
-
- Bernard HU. Coevolution of papillomaviruses with human populations. Trends in microbiology. 1994;2:140–3. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

