Multiple sporadic colorectal cancers display a unique methylation phenotype
- PMID: 24643221
- PMCID: PMC3958343
- DOI: 10.1371/journal.pone.0091033
Multiple sporadic colorectal cancers display a unique methylation phenotype
Abstract
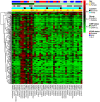
Epigenetics are thought to play a major role in the carcinogenesis of multiple sporadic colorectal cancers (CRC). Previous studies have suggested concordant DNA hypermethylation between tumor pairs. However, only a few methylation markers have been analyzed. This study was aimed at describing the epigenetic signature of multiple CRC using a genome-scale DNA methylation profiling. We analyzed 12 patients with synchronous CRC and 29 age-, sex-, and tumor location-paired patients with solitary tumors from the EPICOLON II cohort. DNA methylation profiling was performed using the Illumina Infinium HM27 DNA methylation assay. The most significant results were validated by Methylight. Tumors samples were also analyzed for the CpG Island Methylator Phenotype (CIMP); KRAS and BRAF mutations and mismatch repair deficiency status. Functional annotation clustering was performed. We identified 102 CpG sites that showed significant DNA hypermethylation in multiple tumors with respect to the solitary counterparts (difference in β value ≥0.1). Methylight assays validated the results for 4 selected genes (p = 0.0002). Eight out of 12(66.6%) multiple tumors were classified as CIMP-high, as compared to 5 out of 29(17.2%) solitary tumors (p = 0.004). Interestingly, 76 out of the 102 (74.5%) hypermethylated CpG sites found in multiple tumors were also seen in CIMP-high tumors. Functional analysis of hypermethylated genes found in multiple tumors showed enrichment of genes involved in different tumorigenic functions. In conclusion, multiple CRC are associated with a distinct methylation phenotype, with a close association between tumor multiplicity and CIMP-high. Our results may be important to unravel the underlying mechanism of tumor multiplicity.
Conflict of interest statement
Figures



References
-
- Chen HS, Sheen-Chen SM (2000) Synchronous and “early” metachronous colorectal adenocarcinoma: analysis of prognosis and current trends. Dis Colon Rectum 43: 1093–1099. - PubMed
-
- Latournerie M, Jooste V, Cottet V, Lepage C, Faivre J, et al. (2008) Epidemiology and prognosis of synchronous colorectal cancers. Br J Surg 95: 1528–1533. - PubMed
-
- Pinol V, Andreu M, Castells A, Paya A, Bessa X, et al. (2004) Synchronous colorectal neoplasms in patients with colorectal cancer: predisposing individual and familial factors. Dis Colon Rectum 47: 1192–1200. - PubMed
-
- Ogino S, Brahmandam M, Kawasaki T, Kirkner GJ, Loda M, et al. (2006) Epigenetic profiling of synchronous colorectal neoplasias by quantitative DNA methylation analysis. Mod Pathol 19: 1083–1090. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

