In search of a candidate pathogen for giant cell arteritis: sequencing-based characterization of the giant cell arteritis microbiome
- PMID: 24644069
- PMCID: PMC4113339
- DOI: 10.1002/art.38631
In search of a candidate pathogen for giant cell arteritis: sequencing-based characterization of the giant cell arteritis microbiome
Abstract
Objective: To characterize the microbiome of the temporal artery in patients with giant cell arteritis (GCA), and to apply an unbiased and comprehensive shotgun sequencing-based approach to determine whether there is an enrichment of candidate pathogens in the affected tissue.
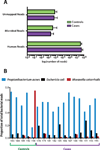
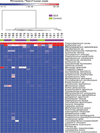
Methods: Temporal artery biopsy specimens were collected from patients at a single institution over a period of 4 years, and unbiased DNA sequencing was performed on 17 formalin-fixed, paraffin-embedded specimens. Twelve of the 17 patients fulfilled the clinical and histopathologic criteria for GCA, and the other 5 patients served as controls. Using PathSeq software, human DNA sequences were computationally subtracted, and the remaining non-human DNA sequences were taxonomically classified using a comprehensive microbial sequence database. The relative abundance of microbes was inferred based on read counts assigned to each organism. Comparison of the microbial diversity between GCA cases and controls was carried out using hierarchical clustering and linear discriminant analysis of effect size.
Results: Propionibacterium acnes and Escherichia coli were the most abundant microorganisms in 16 of the 17 samples, and Moraxella catarrhalis was the most abundant organism in 1 control sample. Pathogens previously described to be correlated with GCA were not differentially abundant in cases compared to controls. There was not a significant burden of likely pathogenic viruses.
Conclusion: DNA sequencing of temporal artery biopsy specimens from GCA cases, in comparison with non-GCA controls, showed no evidence of previously identified candidate GCA pathogens. A single pathogen was not clearly and consistently associated with GCA in this case series.
Copyright © 2014 by the American College of Rheumatology.
Figures
References
-
- Salvarani C, Gabriel SE, O'Fallon WM, Hunder GG. The incidence of giant cell arteritis in Olmsted County, Minnesota: apparent fluctuations in a cyclic pattern. Annals of internal medicine. 1995;123(3):192–194. - PubMed
-
- Gabriel SE, Espy M, Erdman DD, Bjornsson J, Smith TF, Hunder GG. The role of parvovirus B19 in the pathogenesis of giant cell arteritis: a preliminary evaluation. Arthritis and rheumatism. 1999;42(6):1255–1258. - PubMed
-
- Mitchell BM, Font RL. Detection of varicella zoster virus DNA in some patients with giant cell arteritis. Investigative ophthalmology & visual science. 2001;42(11):2572–2577. - PubMed
-
- Haugeberg G, Bie R, Nordbo SA. Temporal arteritis associated with Chlamydia pneumoniae DNA detected in an artery specimen. The Journal of rheumatology. 2001;28(7):1738–1739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

