Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance
- PMID: 24647101
- PMCID: PMC4678626
- DOI: 10.1038/ncomms4446
Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance
Abstract
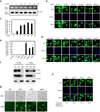
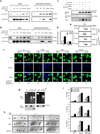
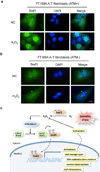
Superoxide dismutase 1 (Sod1) has been known for nearly half a century for catalysis of superoxide to hydrogen peroxide. Here we report a new Sod1 function in oxidative signalling: in response to elevated endogenous and exogenous reactive oxygen species (ROS), Sod1 rapidly relocates into the nucleus, which is important for maintaining genomic stability. Interestingly, H2O2 is sufficient to promote Sod1 nuclear localization, indicating that it is responding to general ROS rather than Sod1 substrate superoxide. ROS signalling is mediated by Mec1/ATM and its effector Dun1/Cds1 kinase, through Dun1 interaction with Sod1 and regulation of Sod1 by phosphorylation at S60, 99. In the nucleus, Sod1 binds to promoters and regulates the expression of oxidative resistance and repair genes. Altogether, our study unravels an unorthodox function of Sod1 as a transcription factor and elucidates the regulatory mechanism for its localization.
Figures



References
-
- Apel K, Hirt H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annual Review of Plant Biology. 2004;55:373–399. - PubMed
-
- COOKE MS, EVANS MD, DIZDAROGLU M, LUNEC J. Oxidative DNA damage: mechanisms, mutation, and disease. The FASEB Journal. 2003;17:1195–1214. - PubMed
-
- Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A Fraction of Yeast Cu,Zn-Superoxide Dismutase and Its Metallochaperone, CCS, Localize to the Intermembrane Space of Mitochondria: A PHYSIOLOGICAL ROLE FOR SOD1 IN GUARDING AGAINST MITOCHONDRIAL OXIDATIVE DAMAGE. Journal of Biological Chemistry. 2001;276:38084–38089. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

