ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays
- PMID: 24647355
- PMCID: PMC3960247
- DOI: 10.1371/journal.pone.0092444
ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays
Abstract
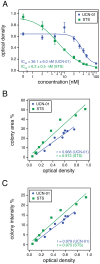
The clonogenic or colony formation assay is a widely used method to study the number and size of cancer cell colonies that remain after irradiation or cytotoxic agent administration and serves as a measure for the anti-proliferative effect of these treatments. Alternatively, this assay is used to quantitate the transforming potential of cancer associated genes and chemical agents. Therefore, there is a need for a simplified and standardized analysis of colony formation assays for both routine laboratory use and for parallelized automated analysis. Here we describe the freely available ImageJ-plugin "ColonyArea", which is optimized for rapid and quantitative analysis of focus formation assays conducted in 6- to 24-well dishes. ColonyArea processes image data of multi-well dishes, by separating, concentrically cropping and background correcting well images individually, before colony formation is quantitated. Instead of counting the number of colonies, ColonyArea determines the percentage of area covered by crystal violet stained cell colonies, also taking the intensity of the staining and therefore cell density into account. We demonstrate that these parameters alone or in combination allow for robust quantification of IC50 values of the cytotoxic effect of two staurosporines, UCN-01 and staurosporine (STS) on human glioblastoma cells (T98G). The relation between the potencies of the two compounds compared very well with that obtained from an absorbance based method to quantify colony growth and to published data. The ColonyArea ImageJ plugin provides a simple and efficient analysis routine to quantitate assay data of one of the most commonly used cellular assays. The bundle is freely available for download as supporting information. We expect that ColonyArea will be of broad utility for cancer biologists, as well as clinical radiation scientists.
Conflict of interest statement
Figures





References
-
- Booden MA, Ulku AS (2005) Cellular Assays of Oncogene. In: Celis JE, editor. Cell Biology, Four-Volume Set, Third Edition: A Laboratory Handbook. Burlington, MA, USA: Elsevier Academic Press, Vol. 3. pp. 345–352.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

