Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv
- PMID: 24647560
- PMCID: PMC3960257
- DOI: 10.1371/journal.pone.0092501
Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv
Erratum in
- PLoS One. 2014;9(6):e101100
Abstract
Background: Antarctic hairgrass (Deschampsia antarctica Desv.) is the only natural grass species in the maritime Antarctic. It has been researched as an important ecological marker and as an extremophile plant for studies on stress tolerance. Despite its importance, little genomic information is available for D. antarctica. Here, we report the complete chloroplast genome, transcriptome profiles of the coding/noncoding genes, and the posttranscriptional processing by RNA editing in the chloroplast system.
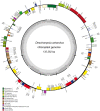
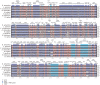
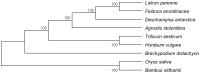
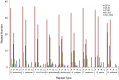
Results: The complete chloroplast genome of D. antarctica is 135,362 bp in length with a typical quadripartite structure, including the large (LSC: 79,881 bp) and small (SSC: 12,519 bp) single-copy regions, separated by a pair of identical inverted repeats (IR: 21,481 bp). It contains 114 unique genes, including 81 unique protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Sequence divergence analysis with other plastomes from the BEP clade of the grass family suggests a sister relationship between D. antarctica, Festuca arundinacea and Lolium perenne of the Poeae tribe, based on the whole plastome. In addition, we conducted high-resolution mapping of the chloroplast-derived transcripts. Thus, we created an expression profile for 81 protein-coding genes and identified ndhC, psbJ, rps19, psaJ, and psbA as the most highly expressed chloroplast genes. Small RNA-seq analysis identified 27 small noncoding RNAs of chloroplast origin that were preferentially located near the 5'- or 3'-ends of genes. We also found >30 RNA-editing sites in the D. antarctica chloroplast genome, with a dominance of C-to-U conversions.
Conclusions: We assembled and characterized the complete chloroplast genome sequence of D. antarctica and investigated the features of the plastid transcriptome. These data may contribute to a better understanding of the evolution of D. antarctica within the Poaceae family for use in molecular phylogenetic studies and may also help researchers understand the characteristics of the chloroplast transcriptome.
Conflict of interest statement
Figures



Similar articles
-
Transcriptome sequencing of the Antarctic vascular plant Deschampsia antarctica Desv. under abiotic stress.Planta. 2013 Mar;237(3):823-36. doi: 10.1007/s00425-012-1797-5. Epub 2012 Nov 8. Planta. 2013. PMID: 23135329
-
The Plastid Genome of Deschampsia cespitosa (Poaceae).Molecules. 2019 Jan 9;24(2):216. doi: 10.3390/molecules24020216. Molecules. 2019. PMID: 30634385 Free PMC article.
-
Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny.BMC Plant Biol. 2020 Sep 2;20(1):406. doi: 10.1186/s12870-020-02621-y. BMC Plant Biol. 2020. PMID: 32878602 Free PMC article.
-
The complete chloroplast genome sequence of the Phyllostachys sulphurea (Poaceae: Bambusoideae).Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(2):983-5. doi: 10.3109/19401736.2014.926516. Epub 2014 Jun 18. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 24938113
-
Introgression mapping in the grasses.Chromosome Res. 2007;15(1):105-13. doi: 10.1007/s10577-006-1103-0. Chromosome Res. 2007. PMID: 17295130 Review.
Cited by
-
Plastome phylogenomics and characterization of rare genomic changes as taxonomic markers in plastome groups 1 and 2 Poeae (Pooideae; Poaceae).PeerJ. 2019 Jun 3;7:e6959. doi: 10.7717/peerj.6959. eCollection 2019. PeerJ. 2019. PMID: 31198631 Free PMC article.
-
The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae.PLoS One. 2018 Apr 25;13(4):e0196069. doi: 10.1371/journal.pone.0196069. eCollection 2018. PLoS One. 2018. PMID: 29694416 Free PMC article.
-
Complete Chloroplast Genomes of Acanthochlamys bracteata (China) and Xerophyta (Africa) (Velloziaceae): Comparative Genomics and Phylogenomic Placement.Front Plant Sci. 2021 Jun 14;12:691833. doi: 10.3389/fpls.2021.691833. eCollection 2021. Front Plant Sci. 2021. PMID: 34194461 Free PMC article.
-
A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions.PeerJ. 2018 Feb 2;6:e4299. doi: 10.7717/peerj.4299. eCollection 2018. PeerJ. 2018. PMID: 29416954 Free PMC article.
-
Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua.Molecules. 2017 Aug 11;22(8):1330. doi: 10.3390/molecules22081330. Molecules. 2017. PMID: 28800082 Free PMC article.
References
-
- Neuhaus HE, Emes MJ (2000) Nonphothosynthetic metabolism in plastids. Annual Review of Plant Physiology and Plant Molecular Biology 51: 111–140. - PubMed
-
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, et al. (2006) the complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution 23: 2175–2190. - PubMed
-
- Bock R (2007) Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids: Springer Berlin Heidelberg. pp. 29–63.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

