Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity
- PMID: 24648345
- PMCID: PMC4012613
- DOI: 10.1158/0008-5472.CAN-13-2482
Comparative functional analysis of DPYD variants of potential clinical relevance to dihydropyrimidine dehydrogenase activity
Abstract
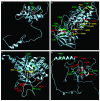
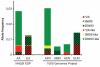
Dihydropyrimidine dehydrogenase (DPD) is the initial and rate-limiting enzyme of the uracil catabolic pathway, being critically important for inactivation of the commonly prescribed anti-cancer drug 5-fluorouracil (5-FU). DPD impairment leads to increased exposure to 5-FU and, in turn, increased anabolism of 5-FU to cytotoxic nucleotides, resulting in more severe clinical adverse effects. Numerous variants within the gene coding for DPD, DPYD, have been described, although only a few have been demonstrated to reduce DPD enzyme activity. To identify DPYD variants that alter enzyme function, we expressed 80 protein-coding variants in an isogenic mammalian system and measured their capacities to convert 5-FU to dihydro-fluorouracil, the product of DPD catabolism. The M166V, E828K, K861R, and P1023T variants exhibited significantly higher enzyme activity than wild-type DPD (120%, P = 0.025; 116%, P = 0.049; 130%, P = 0.0077; 138%, P = 0.048, respectively). Consistent with clinical association studies of 5-FU toxicity, the D949V substitution reduced enzyme activity by 41% (P = 0.0031). Enzyme activity was also significantly reduced for 30 additional variants, 19 of which had <25% activity. None of those 30 variants have been previously reported to associate with 5-FU toxicity in clinical association studies, which have been conducted primarily in populations of European ancestry. Using publicly available genotype databases, we confirmed the rarity of these variants in European populations but showed that they are detected at appreciable frequencies in other populations. These data strongly suggest that testing for the reported deficient DPYD variations could dramatically improve predictive genetic tests for 5-FU sensitivity, especially in individuals of non-European descent.
©2014 AACR.
Figures



References
-
- Meinsma R, Fernandez-Salguero P, Van Kuilenburg AB, Van Gennip AH, Gonzalez FJ. Human polymorphism in drug metabolism: mutation in the dihydropyrimidine dehydrogenase gene results in exon skipping and thymine uracilurea. DNA Cell Biol. 1995;14:1–6. - PubMed
-
- van Kuilenburg AB, Hausler P, Schalhorn A, Tanck MW, Proost JH, Terborg C, et al. Evaluation of 5-fluorouracil pharmacokinetics in cancer patients with a c.1905+1G>A mutation in DPYD by means of a Bayesian limited sampling strategy. Clinical pharmacokinetics. 2012;51:163–74. - PubMed
-
- Johnson MR, Hageboutros A, Wang K, High L, Smith JB, Diasio RB. Life-threatening toxicity in a dihydropyrimidine dehydrogenase-deficient patient after treatment with topical 5-fluorouracil. Clin Cancer Res. 1999;5:2006–11. - PubMed
-
- Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

