Nucleophosmin modulates stability, activity, and nucleolar accumulation of base excision repair proteins
- PMID: 24648491
- PMCID: PMC4019495
- DOI: 10.1091/mbc.E13-12-0717
Nucleophosmin modulates stability, activity, and nucleolar accumulation of base excision repair proteins
Abstract
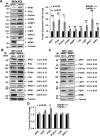
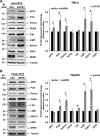
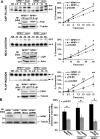
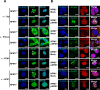
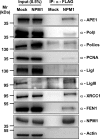
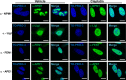
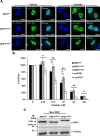
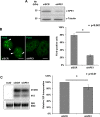
Nucleophosmin (NPM1) is a multifunctional protein that controls cell growth and genome stability via a mechanism that involves nucleolar-cytoplasmic shuttling. It is clear that NPM1 also contributes to the DNA damage response, yet its exact function is poorly understood. We recently linked NPM1 expression to the functional activation of the major abasic endonuclease in mammalian base excision repair (BER), apurinic/apyrimidinic endonuclease 1 (APE1). Here we unveil a novel role for NPM1 as a modulator of the whole BER pathway by 1) controlling BER protein levels, 2) regulating total BER capacity, and 3) modulating the nucleolar localization of several BER enzymes. We find that cell treatment with the genotoxin cisplatin leads to concurrent relocalization of NPM1 and BER components from nucleoli to the nucleoplasm, and cellular experiments targeting APE1 suggest a role for the redistribution of nucleolar BER factors in determining cisplatin toxicity. Finally, based on the use of APE1 as a representative protein of the BER pathway, our data suggest a function for BER proteins in the regulation of ribogenesis.
© 2014 Poletto et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
References
-
- Ali MM, Frei E, Straub J, Breuer A, Wiessler M. Induction of metallothionein by zinc protects from daunorubicin toxicity in rats. Toxicology. 2002;179:85–93. - PubMed
-
- Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet. 2006;15:2239–2249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

