Casein kinase 1δ functions at the centrosome and Golgi to promote ciliogenesis
- PMID: 24648492
- PMCID: PMC4019494
- DOI: 10.1091/mbc.E13-10-0598
Casein kinase 1δ functions at the centrosome and Golgi to promote ciliogenesis
Abstract
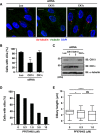
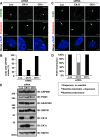
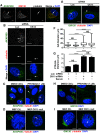
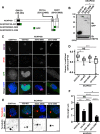
Inhibition of casein kinase 1 delta (CK1δ) blocks primary ciliogenesis in human telomerase reverse transcriptase immortalized retinal pigmented epithelial and mouse inner medullary collecting duct cells-3. Mouse embryonic fibroblasts (MEFs) and retinal cells from Csnk1d (CK1δ)-null mice also exhibit ciliogenesis defects. CK1δ catalytic activity and centrosomal localization signal (CLS) are required to rescue cilia formation in MEFs(Csnk1d null). Furthermore, expression of a truncated derivative containing the CLS displaces full-length CK1δ from the centrosome and decreases ciliary length in control MEFs, suggesting that centrosomal CK1δ has a role in ciliogenesis. CK1δ inhibition also alters pericentrosomal or ciliary distribution of several proteins involved in ciliary transport, including Ras-like in rat brain-11A, Ras-like in rat brain-8A, centrosomal protein of 290 kDa, pericentriolar material protein 1, and polycystin-2, as well as the Golgi distribution of its binding partner, A-kinase anchor protein 450 (AKAP450). As reported for AKAP450, CK1δ was required for microtubule nucleation at the Golgi and maintenance of Golgi integrity. Overexpression of an AKAP450 fragment containing the CK1δ-binding site inhibits Golgi-derived microtubule nucleation, Golgi distribution of intraflagellar transport protein 20 homologue, and ciliogenesis. Our results suggest that CK1δ mediates primary ciliogenesis by multiple mechanisms, one involving its centrosomal function and another dependent on its interaction with AKAP450 at the Golgi, where it is important for maintaining Golgi organization and polarized trafficking of multiple factors that mediate ciliary transport.
© 2014 Greer et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures




References
-
- Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, Knippschild U. Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur J Cell Biol. 2000;79:240–251. - PubMed
-
- Cheong JK, Virshup DM. Casein kinase 1: complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. - PubMed
-
- Coppieters F, Lefever S, Leroy BP, De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat. 2010;31:1097–1108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

