IRF-1 regulates alternative mRNA splicing of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in breast epithelial cells generating an immunoreceptor tyrosine-based inhibition motif (ITIM) containing isoform
- PMID: 24650050
- PMCID: PMC4113144
- DOI: 10.1186/1476-4598-13-64
IRF-1 regulates alternative mRNA splicing of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in breast epithelial cells generating an immunoreceptor tyrosine-based inhibition motif (ITIM) containing isoform
Abstract
Background: Interferon regulatory factor-1 (IRF-1) is a master regulator of IFN-γ induced gene transcription. Previously we have shown that IRF-1 transcriptionally induces CEACAM1 via an ISRE (Interferon-Stimulated Response Element) in its promoter. CEACAM1 pre-mRNA undergoes extensive alternative splicing (AS) generating isoforms to produce either a short (S) cytoplasmic domain expressed primarily in epithelial cells or as an ITIM-containing long (L) isoform in immune cells.
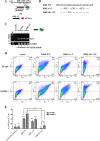
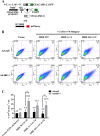
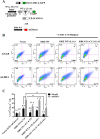
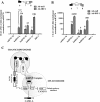
Methods: The transcriptional and molecular mechanism of CEACAM1 minigenes AS containing promoter ISREs mutations in the breast epithelial, MDA-MB-468, cell line was detected using flow cytometry. In addition, transcriptome sequencing was utilized to determine whether IRF-1 could direct the AS of other genes as well. Tumor xenografts were used to evaluate CEACAM1 isoform expression on the leading edge of breast tumor cells.
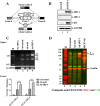
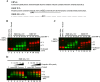
Results: In the present study, we provide evidence that CEACAM1's promoter and variable exon 7 cross-talk allowing IRF-1 to direct AS events. Transcriptome sequencing shows that IRF-1 can also induce the global AS of genes involved in regulation of growth and differentiation as well as genes of the cytokine family. Furthermore, MDA-MB-468 cells grown as tumor xenografts exhibit an AS switch to the L-isoform of CEACAM1, demonstrating that an in vivo inflammatory milieu is also capable of generating the AS switch, similar to that found in human breast cancers Mol Cancer 7:46, 2008.
Conclusions: The novel AS regulatory activities attributed to IRF-1 indicate that the IFN-γ response involves a global change in both gene transcription and AS in breast epithelial cells.
Figures



References
-
- Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

