Relaxed rotational and scrunching changes in P266L mutant of T7 RNA polymerase reduce short abortive RNAs while delaying transition into elongation
- PMID: 24651161
- PMCID: PMC3961267
- DOI: 10.1371/journal.pone.0091859
Relaxed rotational and scrunching changes in P266L mutant of T7 RNA polymerase reduce short abortive RNAs while delaying transition into elongation
Abstract
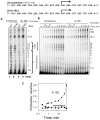
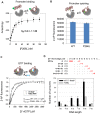
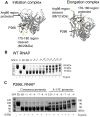
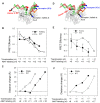
Abortive cycling is a universal feature of transcription initiation catalyzed by DNA-dependent RNA polymerases (RNAP). In bacteriophage T7 RNAP, mutation of proline 266 to leucine (P266L) in the C-linker region connecting the N-terminal promoter binding domain with the C-terminal catalytic domain drastically reduces short abortive products (4-7 nt) while marginally increasing long abortives (9-11 nt). Here we have investigated the transcription initiation pathway of P266L with the goal of understanding the mechanistic basis for short and long abortive synthesis. We show that the P266L mutation does not alter the affinity for the promoter, mildly affects promoter opening, and increases the +1/+2 GTP K(d) by 2-fold. However, unlike wild-type T7 RNAP that undergoes stepwise rotation of the promoter binding domain and DNA scrunching during initial transcription, the P266L mutant does not undergo coupled rotational/scrunching movements until 7 nt RNA synthesis. The lack of rotation/scrunching correlates with greater stabilities of the initiation complexes of the P266L and decreased short abortive products. The results indicate that the increased flexibility in the C-linker due to P266L mutation enables T7 RNAP to absorb the stress from the growing RNA:DNA hybrid thereby decreasing short abortive products. Increased C-linker flexibility, however, has an adverse effect of delaying the transition into elongation by 1-2 nt, which gives rise to long abortive products. However, a mutation in the upstream promoter region greatly decreases long abortive products in P266L reactions, rendering the combination of P266L and A-15C promoter a desirable pair for efficient in vitro transcription for RNA production. We conclude that the conformational rigidity in the C-linker region conferred by the proline at position 266 is responsible for the undesirable short abortive products, but the rigidity is critical for efficient promoter clearance and transition into elongation.
Conflict of interest statement
Figures



Similar articles
-
New insights into the mechanism of initial transcription: the T7 RNA polymerase mutant P266L transitions to elongation at longer RNA lengths than wild type.J Biol Chem. 2012 Oct 26;287(44):37352-61. doi: 10.1074/jbc.M112.370643. Epub 2012 Aug 24. J Biol Chem. 2012. PMID: 22923611 Free PMC article.
-
Sequential release of promoter contacts during transcription initiation to elongation transition.J Mol Biol. 2006 Jul 7;360(2):466-83. doi: 10.1016/j.jmb.2006.05.029. Epub 2006 May 26. J Mol Biol. 2006. PMID: 16780876
-
Transcription initiation in a single-subunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains.Mol Cell. 2008 Jun 6;30(5):567-77. doi: 10.1016/j.molcel.2008.04.003. Mol Cell. 2008. PMID: 18538655 Free PMC article.
-
Snapshots of a viral RNA polymerase switching gears from transcription initiation to elongation.Virol Sin. 2013 Dec;28(6):337-44. doi: 10.1007/s12250-013-3397-3. Epub 2013 Dec 2. Virol Sin. 2013. PMID: 24306760 Free PMC article. Review.
-
The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase.Curr Opin Struct Biol. 2004 Feb;14(1):4-9. doi: 10.1016/j.sbi.2004.01.006. Curr Opin Struct Biol. 2004. PMID: 15102443 Review.
Cited by
-
The In Vivo and In Vitro Architecture of the Hepatitis C Virus RNA Genome Uncovers Functional RNA Secondary and Tertiary Structures.J Virol. 2022 Apr 27;96(8):e0194621. doi: 10.1128/jvi.01946-21. Epub 2022 Mar 30. J Virol. 2022. PMID: 35353000 Free PMC article.
-
Comprehensive evaluation of T7 promoter for enhanced yield and quality in mRNA production.Sci Rep. 2024 Apr 26;14(1):9655. doi: 10.1038/s41598-024-59978-5. Sci Rep. 2024. PMID: 38671016 Free PMC article.
-
Effective synthesis of circRNA via a thermostable T7 RNA polymerase variant as the catalyst.Front Bioeng Biotechnol. 2024 Apr 9;12:1356354. doi: 10.3389/fbioe.2024.1356354. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38655387 Free PMC article.
-
Native Purification and Analysis of Long RNAs.Methods Enzymol. 2015;558:3-37. doi: 10.1016/bs.mie.2015.01.008. Epub 2015 Feb 27. Methods Enzymol. 2015. PMID: 26068736 Free PMC article.
-
An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts.Nat Biotechnol. 2023 Apr;41(4):560-568. doi: 10.1038/s41587-022-01525-6. Epub 2022 Nov 10. Nat Biotechnol. 2023. PMID: 36357718 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

