P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP
- PMID: 24651434
- PMCID: PMC3973206
- DOI: 10.1038/cddis.2014.79
P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP
Abstract
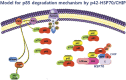
The short isoform of ErbB3-binding protein 1 (Ebp1), p42, is considered to be a potent tumor suppressor in a number of human cancers, although the mechanism by which it exerts this tumor-suppressive activity is unclear. Here, we report that p42 interacts with the cSH2 domain of the p85 subunit of phosphathidyl inositol 3-kinase (PI3K), leading to inhibition of its lipid kinase activity. Importantly, we found that p42 induces protein degradation of the p85 subunit and further identified HSP70/CHIP complex as a novel E3 ligase for p85 that is responsible for p85 ubiquitination and degradation. In this process, p42 couples p85 to the HSP70/CHIP-mediated ubiquitin-proteasomal system (UPS), thereby promoting a reduction of p85 levels both in vitro and in vivo. Thus, the tumor-suppressing effects of p42 in cancer cells are driven by negative regulation of the p85 subunit of PI3K.
Figures





Similar articles
-
C-terminal domain of p42 Ebp1 is essential for down regulation of p85 subunit of PI3K, inhibiting tumor growth.Sci Rep. 2016 Jul 28;6:30626. doi: 10.1038/srep30626. Sci Rep. 2016. PMID: 27464702 Free PMC article.
-
Distinct Interactions of EBP1 Isoforms with FBXW7 Elicits Different Functions in Cancer.Cancer Res. 2017 Apr 15;77(8):1983-1996. doi: 10.1158/0008-5472.CAN-16-2246. Epub 2017 Feb 16. Cancer Res. 2017. PMID: 28209614 Free PMC article.
-
The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity.Oncogene. 2013 Mar 7;32(10):1284-95. doi: 10.1038/onc.2012.144. Epub 2012 Apr 30. Oncogene. 2013. PMID: 22543587
-
Opposing roles of the two isoforms of ErbB3 binding protein 1 in human cancer cells.Int J Cancer. 2016 Sep 15;139(6):1202-8. doi: 10.1002/ijc.30165. Epub 2016 May 17. Int J Cancer. 2016. PMID: 27130196 Review.
-
CHIP: a co-chaperone for degradation by the proteasome.Subcell Biochem. 2015;78:219-42. doi: 10.1007/978-3-319-11731-7_11. Subcell Biochem. 2015. PMID: 25487024 Review.
Cited by
-
Regulation of ribosomal RNA synthesis in T cells: requirement for GTP and Ebp1.Blood. 2015 Apr 16;125(16):2519-29. doi: 10.1182/blood-2014-12-616433. Epub 2015 Feb 17. Blood. 2015. PMID: 25691158 Free PMC article.
-
PA2G4/EBP1 ubiquitination by PRKN/PARKIN promotes mitophagy protecting neuron death in cerebral ischemia.Autophagy. 2024 Feb;20(2):365-379. doi: 10.1080/15548627.2023.2259215. Epub 2023 Sep 15. Autophagy. 2024. PMID: 37712850 Free PMC article.
-
EBP1 promotes the malignant biological behaviors of kidney renal clear cell carcinoma through activation of p38/HIF-1α signaling pathway.Cancer Cell Int. 2024 Jul 24;24(1):261. doi: 10.1186/s12935-024-03442-x. Cancer Cell Int. 2024. PMID: 39049021 Free PMC article.
-
The roles of multifunctional protein ErbB3 binding protein 1 (EBP1) isoforms from development to disease.Exp Mol Med. 2020 Jul;52(7):1039-1047. doi: 10.1038/s12276-020-0476-z. Epub 2020 Jul 27. Exp Mol Med. 2020. PMID: 32719408 Free PMC article. Review.
-
Insulin resistance uncoupled from dyslipidemia due to C-terminal PIK3R1 mutations.JCI Insight. 2016 Oct 20;1(17):e88766. doi: 10.1172/jci.insight.88766. JCI Insight. 2016. PMID: 27766312 Free PMC article.
References
-
- Zhang Y, Hamburger AW. Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription. J Biol Chem. 2004;279:26126–26133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

