High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population
- PMID: 24651479
- PMCID: PMC3961216
- DOI: 10.1371/journal.pone.0090714
High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population
Erratum in
- PLoS One. 2014;9(7):e102939
Abstract
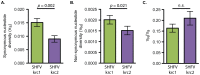
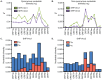
Key biological properties such as high genetic diversity and high evolutionary rate enhance the potential of certain RNA viruses to adapt and emerge. Identifying viruses with these properties in their natural hosts could dramatically improve disease forecasting and surveillance. Recently, we discovered two novel members of the viral family Arteriviridae: simian hemorrhagic fever virus (SHFV)-krc1 and SHFV-krc2, infecting a single wild red colobus (Procolobus rufomitratus tephrosceles) in Kibale National Park, Uganda. Nearly nothing is known about the biological properties of SHFVs in nature, although the SHFV type strain, SHFV-LVR, has caused devastating outbreaks of viral hemorrhagic fever in captive macaques. Here we detected SHFV-krc1 and SHFV-krc2 in 40% and 47% of 60 wild red colobus tested, respectively. We found viral loads in excess of 10(6)-10(7) RNA copies per milliliter of blood plasma for each of these viruses. SHFV-krc1 and SHFV-krc2 also showed high genetic diversity at both the inter- and intra-host levels. Analyses of synonymous and non-synonymous nucleotide diversity across viral genomes revealed patterns suggestive of positive selection in SHFV open reading frames (ORF) 5 (SHFV-krc2 only) and 7 (SHFV-krc1 and SHFV-krc2). Thus, these viruses share several important properties with some of the most rapidly evolving, emergent RNA viruses.
Conflict of interest statement
Figures
References
-
- Grenfell BT, Pybus OG, Gog JR, Wood JLN, Daly JM, et al. (2004) Unifying the epidemiological and evolutionary dynamics of pathogens. Science (New York, NY) 303: 327–332. - PubMed
-
- King DA, Peckham C, Waage JK, Brownlie J, Woolhouse MEJ (2006) Epidemiology. Infectious diseases: preparing for the future. Science (New York, NY) 313: 1392–1393. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

