Selective chemoprecipitation to enrich nitropeptides from complex proteomes for mass-spectrometric analysis
- PMID: 24651500
- PMCID: PMC4594882
- DOI: 10.1038/nprot.2014.052
Selective chemoprecipitation to enrich nitropeptides from complex proteomes for mass-spectrometric analysis
Abstract
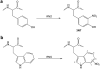
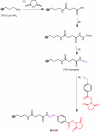
Post-translational protein nitration has attracted interest owing to its involvement in cellular signaling, effects on protein function and potential as biomarker of nitroxidative stress. We describe a procedure for enriching nitropeptides for mass spectrometry (MS)-based proteomics that is a simple and reliable alternative to immunoaffinity-based methods. The starting material for this procedure is a proteolytic digest. The peptides are reacted with formaldehyde and sodium cyanoborohydride to dimethylate all the N-terminal and side chain amino groups. Sodium dithionite is added subsequently to reduce the nitro groups to amines; in theory, the only amino groups present will have originally been nitro groups. The peptide sample is then applied to a solid-phase active ester reagent (SPAER), and those peptides with amino groups will be selectively and covalently captured. Release of the peptides on hydrolysis with trifluoroacetic acid (TFA) results in peptides that have a 4-formyl-benzamido group where the nitro group used to be. In qualitative setups, the procedure can be used to identify proteins modified by reactive nitrogen species and to determine the specific sites of their nitration. Quantitative measurements can be performed by stable-isotope labeling of the peptides in the reductive dimethylation step. Preparation of the SPAER takes about 1 d. Enrichment of nitropeptides requires about 2 d, and sample preparations need 1-30 h, depending on the experimental design. LC-MS/MS assays take from 4 h to several days and data processing can be done in 1-7 d.
Figures


Similar articles
-
Selective chemoprecipitation and subsequent release of tagged species for the analysis of nitropeptides by liquid chromatography-tandem mass spectrometry.Mol Cell Proteomics. 2011 Aug;10(8):M110.002923. doi: 10.1074/mcp.M110.002923. Epub 2011 May 3. Mol Cell Proteomics. 2011. PMID: 21540302 Free PMC article.
-
Relative quantitation of protein nitration by liquid chromatography-mass spectrometry using isotope-coded dimethyl labeling and chemoprecipitation.J Chromatogr A. 2012 Apr 6;1232:266-75. doi: 10.1016/j.chroma.2011.12.100. Epub 2012 Jan 9. J Chromatogr A. 2012. PMID: 22285050 Free PMC article.
-
Quantitative proteomics using reductive dimethylation for stable isotope labeling.J Vis Exp. 2014 Jul 1;(89):51416. doi: 10.3791/51416. J Vis Exp. 2014. PMID: 25045933 Free PMC article.
-
Solid-phase capture for the detection and relative quantification of S-nitrosoproteins by mass spectrometry.Methods. 2013 Aug 1;62(2):130-7. doi: 10.1016/j.ymeth.2012.10.001. Epub 2012 Oct 11. Methods. 2013. PMID: 23064468 Free PMC article. Review.
-
Mass Spectrometry Techniques: Principles and Practices for Quantitative Proteomics.Curr Protein Pept Sci. 2021;22(2):121-133. doi: 10.2174/1389203721666200921153513. Curr Protein Pept Sci. 2021. PMID: 32957902 Review.
Cited by
-
Mass spectrometry-based retina proteomics.Mass Spectrom Rev. 2023 May;42(3):1032-1062. doi: 10.1002/mas.21786. Epub 2022 Jun 6. Mass Spectrom Rev. 2023. PMID: 35670041 Free PMC article. Review.
-
Evaluation of a method for nitrotyrosine site identification and relative quantitation using a stable isotope-labeled nitrated spike-in standard and high resolution fourier transform MS and MS/MS analysis.Int J Mol Sci. 2014 Apr 14;15(4):6265-85. doi: 10.3390/ijms15046265. Int J Mol Sci. 2014. PMID: 24736779 Free PMC article.
-
Uncommon posttranslational modifications in proteomics: ADP-ribosylation, tyrosine nitration, and tyrosine sulfation.Mass Spectrom Rev. 2024 Mar-Apr;43(2):289-326. doi: 10.1002/mas.21811. Epub 2022 Sep 27. Mass Spectrom Rev. 2024. PMID: 36165040 Free PMC article. Review.
-
Direct detection of nitrotyrosine-containing proteins using an aniline-based oxidative coupling strategy.Chem Commun (Camb). 2016 Aug 21;52(65):10036-9. doi: 10.1039/c6cc04575h. Epub 2016 Jul 22. Chem Commun (Camb). 2016. PMID: 27447346 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

