Computational and biological evaluation of N-octadecyl-N'-propylsulfamide, a selective PPARα agonist structurally related to N-acylethanolamines
- PMID: 24651609
- PMCID: PMC3961330
- DOI: 10.1371/journal.pone.0092195
Computational and biological evaluation of N-octadecyl-N'-propylsulfamide, a selective PPARα agonist structurally related to N-acylethanolamines
Abstract
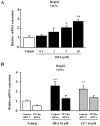
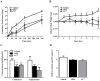
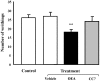
To further understand the pharmacological properties of N-oleoylethanolamine (OEA), a naturally occurring lipid that activates peroxisome proliferator-activated receptor alpha (PPARα), we designed sulfamoyl analogs based on its structure. Among the compounds tested, N-octadecyl-N'-propylsulfamide (CC7) was selected for functional comparison with OEA. The performed studies include the following computational and biological approaches: 1) molecular docking analyses; 2) molecular biology studies with PPARα; 3) pharmacological studies on feeding behavior and visceral analgesia. For the docking studies, we compared OEA and CC7 data with crystallization data obtained with the reference PPARα agonist GW409544. OEA and CC7 interacted with the ligand-binding domain of PPARα in a similar manner to GW409544. Both compounds produced similar transcriptional activation by in vitro assays, including the GST pull-down assay and reporter gene analysis. In addition, CC7 and OEA induced the mRNA expression of CPT1a in HpeG2 cells through PPARα and the induction was avoided with PPARα-specific siRNA. In vivo studies in rats showed that OEA and CC7 had anorectic and antiobesity activity and induced both lipopenia and decreases in hepatic fat content. However, different effects were observed when measuring visceral pain; OEA produced visceral analgesia whereas CC7 showed no effects. These results suggest that OEA activity on the PPARα receptor (e.g., lipid metabolism and feeding behavior) may be dissociated from other actions at alternative targets (e.g., pain) because other non cannabimimetic ligands that interact with PPARα, such as CC7, do not reproduce the full spectrum of the pharmacological activity of OEA. These results provide new opportunities for the development of specific PPARα-activating drugs focused on sulfamide derivatives with a long alkyl chain for the treatment of metabolic dysfunction.
Conflict of interest statement
Figures




Similar articles
-
Elaidyl-sulfamide, an oleoylethanolamide-modelled PPARα agonist, reduces body weight gain and plasma cholesterol in rats.Dis Model Mech. 2012 Sep;5(5):660-70. doi: 10.1242/dmm.009233. Epub 2012 Jun 26. Dis Model Mech. 2012. PMID: 22736460 Free PMC article.
-
Novel sulfamide analogs of oleoylethanolamide showing in vivo satiety inducing actions and PPARalpha activation.J Med Chem. 2007 Jan 25;50(2):389-93. doi: 10.1021/jm0601102. J Med Chem. 2007. PMID: 17228882
-
Identification and characterization of phytocannabinoids as novel dual PPARα/γ agonists by a computational and in vitro experimental approach.Biochim Biophys Acta Gen Subj. 2019 Mar;1863(3):586-597. doi: 10.1016/j.bbagen.2019.01.002. Epub 2019 Jan 3. Biochim Biophys Acta Gen Subj. 2019. PMID: 30611848
-
Neuroprotective properties of peroxisome proliferator-activated receptor alpha (PPARα) and its lipid ligands.Curr Med Chem. 2014;21(24):2803-21. doi: 10.2174/0929867321666140303143455. Curr Med Chem. 2014. PMID: 24606520 Review.
-
Oleoylethanolamide: The role of a bioactive lipid amide in modulating eating behaviour.Obes Rev. 2018 Feb;19(2):178-197. doi: 10.1111/obr.12630. Epub 2017 Nov 10. Obes Rev. 2018. PMID: 29124885 Review.
Cited by
-
Role of the satiety factor oleoylethanolamide in alcoholism.Addict Biol. 2016 Jul;21(4):859-72. doi: 10.1111/adb.12276. Epub 2015 Jun 2. Addict Biol. 2016. PMID: 26037332 Free PMC article.
-
Octadecylpropyl Sulfamide Reduces Neurodegeneration and Restores the Memory Deficits Induced by Hypoxia-Ischemia in Mice.Front Pharmacol. 2018 Apr 19;9:376. doi: 10.3389/fphar.2018.00376. eCollection 2018. Front Pharmacol. 2018. PMID: 29725299 Free PMC article.
-
The endocannabinoid system - a target for the treatment of LUTS?Nat Rev Urol. 2016 Aug;13(8):463-70. doi: 10.1038/nrurol.2016.110. Epub 2016 Jul 5. Nat Rev Urol. 2016. PMID: 27377161 Review.
References
-
- Staels B, Maes M, Zambon A (2008) Fibrates and future PPARalpha agonists in the treatment of cardiovascular disease. Nat Clin Pract Cardiovasc Med 5: 542–553. - PubMed
-
- Willson TM, Brown PJ, Sternbach DD, Henke BR (2000) The PPARs: from orphan receptors to drug discovery. J Med Chem 43: 527–550. - PubMed
-
- Kostadinova R, Wahli W, Michalik L (2005) PPARs in diseases: control mechanisms of inflammation. Curr Med Chem 12: 2995–3009. - PubMed
-
- Singh JP, Kauffman R, Bensch W, Wang G, McClelland P, et al. (2005) Identification of a novel selective peroxisome proliferator-activated receptor alpha agonist, 2-methyl-2-(4-{3-[1-(4-methylbenzyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3- yl]propyl}phenoxy)propanoic acid (LY518674), that produces marked changes in serum lipids and apolipoprotein A-1 expression. Mol Pharmacol 68: 763–768. - PubMed
-
- Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, et al. (2003) Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 425: 90–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

