Contrasting patterns in mammal-bacteria coevolution: bartonella and leptospira in bats and rodents
- PMID: 24651646
- PMCID: PMC3961187
- DOI: 10.1371/journal.pntd.0002738
Contrasting patterns in mammal-bacteria coevolution: bartonella and leptospira in bats and rodents
Abstract
Background: Emerging bacterial zoonoses in bats and rodents remain relatively understudied. We conduct the first comparative host-pathogen coevolutionary analyses of bacterial pathogens in these hosts, using Bartonella spp. and Leptospira spp. as a model.
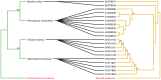
Methodology/principal findings: We used published genetic data for 51 Bartonella genotypes from 24 bat species, 129 Bartonella from 38 rodents, and 26 Leptospira from 20 bats. We generated maximum likelihood and Bayesian phylogenies for hosts and bacteria, and tested for coevoutionary congruence using programs ParaFit, PACO, and Jane. Bartonella spp. and their bat hosts had a significant coevolutionary fit (ParaFitGlobal = 1.9703, P≤0.001; m2 global value = 7.3320, P≤0.0001). Bartonella spp. and rodent hosts also indicated strong overall patterns of cospeciation (ParaFitGlobal = 102.4409, P≤0.001; m2 global value = 86.532, P≤0.0001). In contrast, we were unable to reject independence of speciation events in Leptospira and bats (ParaFitGlobal = 0.0042, P = 0.84; m2 global value = 4.6310, P = 0.5629). Separate analyses of New World and Old World data subsets yielded results congruent with analysis from entire datasets. We also conducted event-based cophylogeny analyses to reconstruct likely evolutionary histories for each group of pathogens and hosts. Leptospira and bats had the greatest number of host switches per parasite (0.731), while Bartonella and rodents had the fewest (0.264).
Conclusions/significance: In both bat and rodent hosts, Bartonella exhibits significant coevolution with minimal host switching, while Leptospira in bats lacks evolutionary congruence with its host and has high number of host switches. Reasons underlying these variable coevolutionary patterns in host range are likely due to differences in disease-specific transmission and host ecology. Understanding the coevolutionary patterns and frequency of host-switching events between bacterial pathogens and their hosts will allow better prediction of spillover between mammal reservoirs, and ultimately to humans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Simmons NB, Wilson D, Reeder D (2005) Order chiroptera. Mammal species of the world: a taxonomic and geographic reference 1: 312–529.
-
- Pagel MD, May RM, Collie AR (1991) Ecological aspects of the geographical distribution and diversity of mammalian species. American Naturalist 137: 791–815.
-
- Olival KJ, Epstein JH, Wang LF, Field HE, Daszak P (2012) Are bats unique viral reservoirs? In: Aguirre AA, Ostfeld RS, Daszak P, editors. New Directions in Conservation Medicine: Applied Cases of Ecological Health. 2nd ed. Oxford: Oxford University Press. pp. 195–212.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

