TALENs mediate efficient and heritable mutation of endogenous genes in the marine annelid Platynereis dumerilii
- PMID: 24653002
- PMCID: PMC4012502
- DOI: 10.1534/genetics.113.161091
TALENs mediate efficient and heritable mutation of endogenous genes in the marine annelid Platynereis dumerilii
Abstract
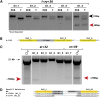
Platynereis dumerilii is a marine polychaete and an established model system for studies of evolution and development. Platynereis is also a re-emerging model for studying the molecular basis of circalunar reproductive timing: a biological phenomenon observed in many marine species. While gene expression studies have provided new insight into patterns of gene regulation, a lack of reverse genetic tools has so far limited the depth of functional analyses in this species. To address this need, we established customized transcriptional activator-like effector nucleases (TALENs) as a tool to engineer targeted modifications in Platynereis genes. By adapting a workflow of TALEN construction protocols and mutation screening approaches for use in Platynereis, we engineered frameshift mutations in three endogenous Platynereis genes. We confirmed that such mutations are heritable, demonstrating that TALENs can be used to generate homozygous knockout lines in P. dumerilii. This is the first use of TALENs for generating genetic knockout mutations in an annelid model. These tools not only open the door for detailed in vivo functional analyses, but also can facilitate further technical development, such as targeted genome editing.
Keywords: chronobiology; evolution; genome editing; invertebrate; marine.
Figures



References
-
- Ansai S., Inohaya K., Yoshiura Y., Schartl M., Uemura N., et al. , 2014. Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka. Dev. Growth Differ. 56: 98–107. - PubMed
-
- Arendt D., Nübler-Jung K., 1994. Inversion of dorsoventral axis? Nature 371: 26. - PubMed
-
- Arendt D., Nübler-Jung K., 1997. Dorsal or ventral: similarities in fate maps and gastrulation patterns in annelids, arthropods and chordates. Mech. Dev. 61: 7–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

