A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma
- PMID: 24654231
- PMCID: PMC4084668
- DOI: 10.1158/0008-5472.CAN-13-2775
A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma
Abstract
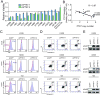
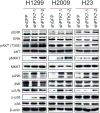
Lung cancer remains the most common cause of cancer-related death worldwide and it continues to lack effective treatment. The increasingly large and diverse public databases of lung cancer gene expression constitute a rich source of candidate oncogenic drivers and therapeutic targets. To define novel targets for lung adenocarcinoma, we conducted a large-scale meta-analysis of genes specifically overexpressed in adenocarcinoma. We identified an 11-gene signature that was overexpressed consistently in adenocarcinoma specimens relative to normal lung tissue. Six genes in this signature were specifically overexpressed in adenocarcinoma relative to other subtypes of non-small cell lung cancer (NSCLC). Among these genes was the little studied protein tyrosine kinase PTK7. Immunohistochemical analysis confirmed that PTK7 is highly expressed in primary adenocarcinoma patient samples. RNA interference-mediated attenuation of PTK7 decreased cell viability and increased apoptosis in a subset of adenocarcinoma cell lines. Further, loss of PTK7 activated the MKK7-JNK stress response pathway and impaired tumor growth in xenotransplantation assays. Our work defines PTK7 as a highly and specifically expressed gene in adenocarcinoma and a potential therapeutic target in this subset of NSCLC.
©2014 American Association for Cancer Research.
Conflict of interest statement
Declaration of conflict of interest: the authors declare no conflict of interest.
Figures




References
-
- Howlader NNA, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975-2009 (Vintage 2009 Populations) [accessed August 13, 2012]; http://seer.cancer.gov/csr/1975_2009_pops09/index.html.
-
- Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:2121–2127. - PubMed
-
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. - PubMed
-
- Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nature medicine. 2012;18:349–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

