pH-dependent activities and structural stability of loop-2-anchoring helix of RadA recombinase from Methanococcus voltae
- PMID: 24654848
- PMCID: PMC4150490
- DOI: 10.2174/0929866521666140320103512
pH-dependent activities and structural stability of loop-2-anchoring helix of RadA recombinase from Methanococcus voltae
Abstract
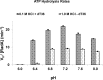
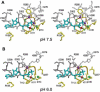
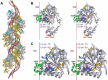
RadA is an archaeal orthologue of human recombinase Rad51. This superfamily of recombinases, which also includes eukaryal meiosis-specific DMC1 and remotely related bacterial RecA, form filaments on single-stranded DNA in the presence of ATP and promote a strand exchange reaction between the single-stranded DNA and a homologous double stranded DNA. Due to its feasibility of getting crystals and similarity (> 40% sequence identity) to eukaryal homologues, we have studied RadA from Methanococcus voltae (MvRadA) as a structural model for understanding the molecular mechanism of homologous strand exchange. Here we show this protein's ATPase and strand exchange activities are minimal at pH 6.0. Interestingly, MvRadA's pH dependence is similar to the properties of human Rad51 but dissimilar to that of the well-studied E. coli RecA. A structure subsequently determined at pH 6.0 reveals features indicative of an ATPase- inactive form with a disordered L2 loop. Comparison with a previously determined ATPase-active form at pH 7.5 implies that the stability of the ATPase-active conformation is reduced at the acidic pH. We interpret these results as further suggesting an ordered disposition of the DNA-binding L2 region, similar to what has been observed in the previously observed ATPase-active conformation, is required for promoting hydrolysis of ATP and strand exchange between singleand double-stranded DNA. His-276 in the mobile L2 region was observed to be partially responsible for the pH-dependent activities of MvRadA.
Figures


Similar articles
-
Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence.J Biol Chem. 2005 Jan 7;280(1):722-8. doi: 10.1074/jbc.M411093200. Epub 2004 Nov 10. J Biol Chem. 2005. PMID: 15537659
-
Binding of a second magnesium is required for ATPase activity of RadA from Methanococcus voltae.Biochemistry. 2007 May 22;46(20):5855-63. doi: 10.1021/bi6024098. Epub 2007 Apr 25. Biochemistry. 2007. PMID: 17455906
-
Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.J Mol Biol. 2006 Jul 14;360(3):537-47. doi: 10.1016/j.jmb.2006.05.058. Epub 2006 Jun 9. J Mol Biol. 2006. PMID: 16782126
-
Structure/function relationships in RecA protein-mediated homology recognition and strand exchange.Crit Rev Biochem Mol Biol. 2015;50(6):453-76. doi: 10.3109/10409238.2015.1092943. Epub 2015 Oct 13. Crit Rev Biochem Mol Biol. 2015. PMID: 26459995 Review.
-
Right or left turn? RecA family protein filaments promote homologous recombination through clockwise axial rotation.Bioessays. 2008 Jan;30(1):48-56. doi: 10.1002/bies.20694. Bioessays. 2008. PMID: 18081011 Review.
References
-
- Cox MM. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3(2):65–78. - PubMed
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404(6773):37–41. - PubMed
-
- Courcelle J, Ganesan AK, Hanawalt PC. Therefore, what are recombination proteins there for?. Bioessays. 2001;23(5):463–70. - PubMed
-
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. - PubMed
-
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25(4):156–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
