Chemical exchange in biomacromolecules: past, present, and future
- PMID: 24656076
- PMCID: PMC4049312
- DOI: 10.1016/j.jmr.2014.01.008
Chemical exchange in biomacromolecules: past, present, and future
Abstract
The perspective reviews quantitative investigations of chemical exchange phenomena in proteins and other biological macromolecules using NMR spectroscopy, particularly relaxation dispersion methods. The emphasis is on techniques and applications that quantify the populations, interconversion kinetics, and structural features of sparsely populated conformational states in equilibrium with a highly populated ground state. Applications to folding, molecular recognition, catalysis, and allostery by proteins and nucleic acids are highlighted.
Keywords: Dynamics; Kinetics; NMR; Nucleic acids; Proteins; Relaxation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

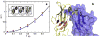

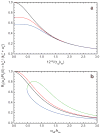





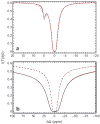
References
-
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. second ed. Academic Press; San Diego, CA: 2007.
-
- Forsén S, Hoffman RA. Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J Chem Phys. 1963;39:2892.
-
- Gutowsky HS, Saika A. Dissociation, chemical exchange, and the proton magnetic resonance in some aqueous electrolytes. J Chem Phys. 1953;21:1688–1694.
-
- Fejzo J, Westler WM, Macura S, Markley JL. Elimination of cross-relaxation effects from two-dimensional chemical-exchange spectra of macromolecules. J Am Chem Soc. 1990;112:2574–2577.
-
- Palmer AG, Rance M, Wright PE. Intramolecular motions of a zinc finger DNA-binding domain from xfin characterized by proton-detected natural abundance 13C heteronuclear NMR spectroscopy. J Am Chem Soc. 1991;113:4371–4380.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

