Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity
- PMID: 24656819
- PMCID: PMC3989773
- DOI: 10.1016/j.celrep.2014.02.019
Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity
Abstract
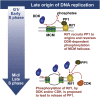
The firing of eukaryotic origins of DNA replication requires CDK and DDK kinase activities. DDK, in particular, is involved in setting the temporal program of origin activation, a conserved feature of eukaryotes. Rif1, originally identified as a telomeric protein, was recently implicated in specifying replication timing in yeast and mammals. We show that this function of Rif1 depends on its interaction with PP1 phosphatases. Mutations of two PP1 docking motifs in Rif1 lead to early replication of telomeres in budding yeast and misregulation of origin firing in fission yeast. Several lines of evidence indicate that Rif1/PP1 counteract DDK activity on the replicative MCM helicase. Our data suggest that the PP1/Rif1 interaction is downregulated by the phosphorylation of Rif1, most likely by CDK/DDK. These findings elucidate the mechanism of action of Rif1 in the control of DNA replication and demonstrate a role of PP1 phosphatases in the regulation of origin firing.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Alvarez-Tabarés I., Grallert A., Ortiz J.M., Hagan I.M. Schizosaccharomyces pombe protein phosphatase 1 in mitosis, endocytosis and a partnership with Wsh3/Tea4 to control polarised growth. J. Cell Sci. 2007;120:3589–3601. - PubMed
-
- Bianchi A., Shore D. Early replication of short telomeres in budding yeast. Cell. 2007;128:1051–1062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

