Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity
- PMID: 24656864
- PMCID: PMC3980428
- DOI: 10.1016/j.ajhg.2014.02.013
Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity
Abstract
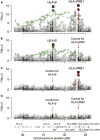
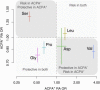
Despite progress in defining human leukocyte antigen (HLA) alleles for anti-citrullinated-protein-autoantibody-positive (ACPA(+)) rheumatoid arthritis (RA), identifying HLA alleles for ACPA-negative (ACPA(-)) RA has been challenging because of clinical heterogeneity within clinical cohorts. We imputed 8,961 classical HLA alleles, amino acids, and SNPs from Immunochip data in a discovery set of 2,406 ACPA(-) RA case and 13,930 control individuals. We developed a statistical approach to identify and adjust for clinical heterogeneity within ACPA(-) RA and observed independent associations for serine and leucine at position 11 in HLA-DRβ1 (p = 1.4 × 10(-13), odds ratio [OR] = 1.30) and for aspartate at position 9 in HLA-B (p = 2.7 × 10(-12), OR = 1.39) within the peptide binding grooves. These amino acid positions induced associations at HLA-DRB1(∗)03 (encoding serine at 11) and HLA-B(∗)08 (encoding aspartate at 9). We validated these findings in an independent set of 427 ACPA(-) case subjects, carefully phenotyped with a highly sensitive ACPA assay, and 1,691 control subjects (HLA-DRβ1 Ser11+Leu11: p = 5.8 × 10(-4), OR = 1.28; HLA-B Asp9: p = 2.6 × 10(-3), OR = 1.34). Although both amino acid sites drove risk of ACPA(+) and ACPA(-) disease, the effects of individual residues at HLA-DRβ1 position 11 were distinct (p < 2.9 × 10(-107)). We also identified an association with ACPA(+) RA at HLA-A position 77 (p = 2.7 × 10(-8), OR = 0.85) in 7,279 ACPA(+) RA case and 15,870 control subjects. These results contribute to mounting evidence that ACPA(+) and ACPA(-) RA are genetically distinct and potentially have separate autoantigens contributing to pathogenesis. We expect that our approach might have broad applications in analyzing clinical conditions with heterogeneity at both major histocompatibility complex (MHC) and non-MHC regions.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Daha N.A., Toes R.E.M. Rheumatoid arthritis: Are ACPA-positive and ACPA-negative RA the same disease? Nat. Rev. Rheumatol. 2011;7:202–203. - PubMed
-
- Eyre S., Bowes J., Diogo D., Lee A., Barton A., Martin P., Zhernakova A., Stahl E., Viatte S., McAllister K., Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate. Wellcome Trust Case Control Consortium High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 2012;44:1336–1340. - PMC - PubMed
-
- Ding B., Padyukov L., Lundström E., Seielstad M., Plenge R.M., Oksenberg J.R., Gregersen P.K., Alfredsson L., Klareskog L. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

