Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes
- PMID: 24657167
- PMCID: PMC3988258
- DOI: 10.1016/j.molcel.2014.02.018
Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes
Abstract
NLR (nucleotide-binding domain [NBD]- and leucine-rich repeat [LRR]-containing) proteins mediate innate immune sensing of pathogens in mammals and plants. How NLRs detect their cognate stimuli remains poorly understood. Here, we analyzed ligand recognition by NLR apoptosis inhibitory protein (NAIP) inflammasomes. Mice express multiple highly related NAIP paralogs that recognize distinct bacterial proteins. We analyzed a panel of 43 chimeric NAIPs, allowing us to map the NAIP domain responsible for specific ligand detection. Surprisingly, ligand specificity was mediated not by the LRR domain, but by an internal region encompassing several NBD-associated α-helical domains. Interestingly, we find that the ligand specificity domain has evolved under positive selection in both rodents and primates. We further show that ligand binding is required for the subsequent co-oligomerization of NAIPs with the downstream signaling adaptor NLR family, CARD-containing 4 (NLRC4). These data provide a molecular basis for how NLRs detect ligands and assemble into inflammasomes.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
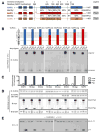
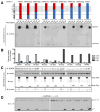
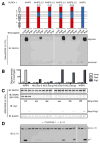
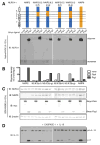

Comment in
-
NAIP inflammasomes give the NOD to bacterial ligands.Trends Immunol. 2014 Nov;35(11):503-4. doi: 10.1016/j.it.2014.10.002. Epub 2014 Oct 18. Trends Immunol. 2014. PMID: 25443492 Free PMC article.
References
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. - PubMed
-
- Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15:763–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

