Breast cancer patient stratification using a molecular regularized consensus clustering method
- PMID: 24657666
- PMCID: PMC4151565
- DOI: 10.1016/j.ymeth.2014.03.005
Breast cancer patient stratification using a molecular regularized consensus clustering method
Abstract
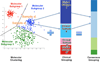
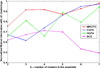
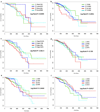
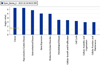
Breast cancers are highly heterogeneous with different subtypes that lead to different clinical outcomes including prognosis, response to treatment and chances of recurrence and metastasis. An important task in personalized medicine is to determine the subtype for a breast cancer patient in order to provide the most effective treatment. In order to achieve this goal, integrative genomics approach has been developed recently with multiple modalities of large datasets ranging from genotypes to multiple levels of phenotypes. A major challenge in integrative genomics is how to effectively integrate multiple modalities of data to stratify the breast cancer patients. Consensus clustering algorithms have often been adopted for this purpose. However, existing consensus clustering algorithms are not suitable for the situation of integrating clustering results obtained from a mixture of numerical data and categorical data. In this work, we present a mathematical formulation for integrative clustering of multiple-source data including both numerical and categorical data to resolve the above issue. Specifically, we formulate the problem as a novel consensus clustering method called Molecular Regularized Consensus Patient Stratification (MRCPS) based on an optimization process with regularization. Unlike the traditional consensus clustering methods, MRCPS can automatically and spontaneously cluster both numerical and categorical data with any option of similarity metrics. We apply this new method by applying it on the TCGA breast cancer datasets and evaluate using both statistical criteria and clinical relevance on predicting prognosis. The result demonstrates the superiority of this method in terms of effectiveness of aggregation and differentiating patient outcomes. Our method, while motivated by the breast cancer research, is nevertheless universal for integrative genomics studies.
Keywords: Breast cancer prognosis; Breast cancer subtypes; Cancer patient stratification; Consensus clustering; Integrative genomic.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
-
- Perou CM, Sø rlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees Ca, Pollack JR, Ross DT, Johnsen H, Akslen La, Fluge O, Pergamenschikov a, Williams C, Zhu SX, Lø nning PE, Bø rresen Dale aL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3465532\&tool=.... - PubMed
-
- van’t Veer L, Dai H, Vijver MVD. Gene expression profiling predicts clinical outcome of breast cancer. nature. 415(345) URL http://www.nature.com/nature/journal/v415/n6871/abs/415530a.html. - PubMed
-
- Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, Huse J, Ladanyi M, Sander C. Integrative subtype discovery in glioblastoma using iCluster. PloS one. 2012;7(4):e35236. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3335101\&tool=.... - PMC - PubMed
-
- Gade S, Porzelius C, Fälth M, Brase JC, Wuttig D, Kuner R, Binder H, Sültmann H, Beissbarth T. Graph based fusion of miRNA and mRNA expression data improves clinical outcome prediction in prostate cancer. BMC bioinformatics. 2011;12:488. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3471479\&tool=.... - PMC - PubMed
-
- Yuan Y, Savage RS, Markowetz F. Patient-specific data fusion defines prognostic cancer subtypes. PLoS computational biology. 2011;7(10):e1002227. URL http://dx.plos.org/10.1371/journal.pcbi.1002227. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

