Draft genome comparison of representatives of the three dominant genotype groups of dairy Bacillus licheniformis strains
- PMID: 24657871
- PMCID: PMC4018868
- DOI: 10.1128/AEM.00065-14
Draft genome comparison of representatives of the three dominant genotype groups of dairy Bacillus licheniformis strains
Abstract
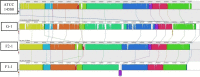
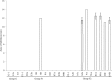
The spore-forming bacterium Bacillus licheniformis is a common contaminant of milk and milk products. Strains of this species isolated from dairy products can be differentiated into three major groups, namely, G, F1, and F2, using random amplification of polymorphic DNA (RAPD) analysis; however, little is known about the genomic differences between these groups and the identity of the fragments that make up their RAPD profiles. In this work we obtained high-quality draft genomes of representative strains from each of the three RAPD groups (designated strain G-1, strain F1-1, and strain F2-1) and compared them to each other and to B. licheniformis ATCC 14580 and Bacillus subtilis 168. Whole-genome comparison and multilocus sequence typing revealed that strain G-1 contains significant sequence variability and belongs to a lineage distinct from the group F strains. Strain G-1 was found to contain genes coding for a type I restriction modification system, urease production, and bacitracin synthesis, as well as the 8-kbp plasmid pFL7, and these genes were not present in strains F1-1 and F2-1. In agreement with this, all isolates of group G, but no group F isolates, were found to possess urease activity and antimicrobial activity against Micrococcus. Identification of RAPD band sequences revealed that differences in the RAPD profiles were due to differences in gene lengths, 3' ends of predicted primer binding sites, or gene presence or absence. This work provides a greater understanding of the phylogenetic and phenotypic differences observed within the B. licheniformis species.
Figures




Similar articles
-
RAPD-based screening for spore-forming bacterial populations in Uruguayan commercial powdered milk.Int J Food Microbiol. 2011 Jul 15;148(1):36-41. doi: 10.1016/j.ijfoodmicro.2011.04.020. Epub 2011 Apr 28. Int J Food Microbiol. 2011. PMID: 21565415
-
A RAPD-based comparison of thermophilic bacilli from milk powders.Int J Food Microbiol. 2003 Aug 15;85(1-2):45-61. doi: 10.1016/s0168-1605(02)00480-4. Int J Food Microbiol. 2003. PMID: 12810270
-
Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7T and FZB42T: a proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons.Int J Syst Evol Microbiol. 2011 Aug;61(Pt 8):1786-1801. doi: 10.1099/ijs.0.023267-0. Epub 2010 Sep 3. Int J Syst Evol Microbiol. 2011. PMID: 20817842
-
Diversity and functionality of Bacillus and related genera isolated from spontaneously fermented soybeans (Indian Kinema) and locust beans (African Soumbala).Int J Food Microbiol. 2002 Aug 25;77(3):175-86. doi: 10.1016/s0168-1605(02)00124-1. Int J Food Microbiol. 2002. PMID: 12160077
-
Development of a RAPD-PCR method for identification of Bacillus species isolated from Cheonggukjang.Int J Food Microbiol. 2009 Feb 28;129(3):282-7. doi: 10.1016/j.ijfoodmicro.2008.12.013. Epub 2008 Dec 24. Int J Food Microbiol. 2009. PMID: 19157616
Cited by
-
Screening and characterization of RAPD markers in viscerotropic Leishmania parasites.PLoS One. 2014 Oct 14;9(10):e109773. doi: 10.1371/journal.pone.0109773. eCollection 2014. PLoS One. 2014. PMID: 25313833 Free PMC article.
-
BsuMI regulates DNA transformation in Bacillus subtilis besides the defense system and the constructed strain with BsuMI-absence is applicable as a universal transformation platform for wild-type Bacillus.Microb Cell Fact. 2024 Aug 9;23(1):225. doi: 10.1186/s12934-024-02493-z. Microb Cell Fact. 2024. PMID: 39123211 Free PMC article.
-
The Prevalence and Control of Bacillus and Related Spore-Forming Bacteria in the Dairy Industry.Front Microbiol. 2015 Dec 21;6:1418. doi: 10.3389/fmicb.2015.01418. eCollection 2015. Front Microbiol. 2015. PMID: 26733963 Free PMC article. Review.
-
Whole genome shotgun sequence of Bacillus paralicheniformis strain KMS 80, a rhizobacterial endophyte isolated from rice (Oryza sativa L.).3 Biotech. 2018 May;8(5):223. doi: 10.1007/s13205-018-1242-y. Epub 2018 Apr 20. 3 Biotech. 2018. PMID: 29692960 Free PMC article.
-
Recent Advancements in the Technologies Detecting Food Spoiling Agents.J Funct Biomater. 2021 Nov 27;12(4):67. doi: 10.3390/jfb12040067. J Funct Biomater. 2021. PMID: 34940546 Free PMC article. Review.
References
-
- Williams DJ. 1958. Bacillus spores in raw milk in Victoria and New South Wales. Aust. J. Dairy Technol. 13:3–10
-
- Cook GM, Sandeman RM. 2000. Sources and characterisation of spore-forming bacteria in raw milk. Aust. J. Dairy Technol 55:119–126
-
- Chauhan K, Dhakal R, Seale RB, Deeth HC, Pillidge CJ, Powell IB, Craven H, Turner MS. 2013. Rapid identification of dairy mesophilic and thermophilic sporeforming bacteria using DNA high resolution melt analysis of variable 16S rDNA regions. Int. J. Food Microbiol. 165:175–183. 10.1016/j.ijfoodmicro.2013.05.007 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

