Extensive microbial and functional diversity within the chicken cecal microbiome
- PMID: 24657972
- PMCID: PMC3962364
- DOI: 10.1371/journal.pone.0091941
Extensive microbial and functional diversity within the chicken cecal microbiome
Abstract
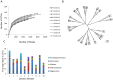
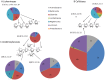
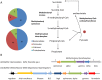
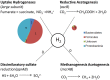
Chickens are major source of food and protein worldwide. Feed conversion and the health of chickens relies on the largely unexplored complex microbial community that inhabits the chicken gut, including the ceca. We have carried out deep microbial community profiling of the microbiota in twenty cecal samples via 16S rRNA gene sequences and an in-depth metagenomics analysis of a single cecal microbiota. We recovered 699 phylotypes, over half of which appear to represent previously unknown species. We obtained 648,251 environmental gene tags (EGTs), the majority of which represent new species. These were binned into over two-dozen draft genomes, which included Campylobacter jejuni and Helicobacter pullorum. We found numerous polysaccharide- and oligosaccharide-degrading enzymes encoding within the metagenome, some of which appeared to be part of polysaccharide utilization systems with genetic evidence for the co-ordination of polysaccharide degradation with sugar transport and utilization. The cecal metagenome encodes several fermentation pathways leading to the production of short-chain fatty acids, including some with novel features. We found a dozen uptake hydrogenases encoded in the metagenome and speculate that these provide major hydrogen sinks within this microbial community and might explain the high abundance of several genera within this microbiome, including Campylobacter, Helicobacter and Megamonas.
Conflict of interest statement
Figures


References
-
- Harre R (1981) Aristotle: the embryology of the chick. Great scientific experiments Oxford: Phaidon. 31–38.
-
- Darwin C (1868) The variation of animals and plants under domestication. London: Murray.
-
- Burt DW (2007) Emergence of the chicken as a model organism: implications for agriculture and biology. Poult Sci 86: 1460–1471. - PubMed
-
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, et al. (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

