Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6
- PMID: 24657997
- PMCID: PMC3962378
- DOI: 10.1371/journal.pone.0092249
Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6
Abstract
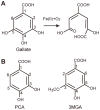
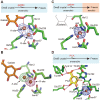
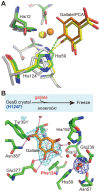
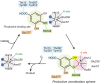
DesB, which is derived from Sphingobium sp. SYK-6, is a type II extradiol dioxygenase that catalyzes a ring opening reaction of gallate. While typical extradiol dioxygenases show broad substrate specificity, DesB has strict substrate specificity for gallate. The substrate specificity of DesB seems to be required for the efficient growth of S. sp. SYK-6 using lignin-derived aromatic compounds. Since direct coordination of hydroxyl groups of the substrate to the non-heme iron in the active site is a critical step for the catalytic reaction of the extradiol dioxygenases, the mechanism of the substrate recognition and coordination of DesB was analyzed by biochemical and crystallographic methods. Our study demonstrated that the direct coordination between the non-heme iron and hydroxyl groups of the substrate requires a large shift of the Fe (II) ion in the active site. Mutational analysis revealed that His124 and His192 in the active site are essential to the catalytic reaction of DesB. His124, which interacts with OH (4) of the bound gallate, seems to contribute to proper positioning of the substrate in the active site. His192, which is located close to OH (3) of the gallate, is likely to serve as the catalytic base. Glu377' interacts with OH (5) of the gallate and seems to play a critical role in the substrate specificity. Our biochemical and structural study showed the substrate recognition and catalytic mechanisms of DesB.
Conflict of interest statement
Figures
Similar articles
-
Crystallization and preliminary crystallographic analysis of gallate dioxygenase DesB from Sphingobium sp. SYK-6.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Nov 1;65(Pt 11):1171-4. doi: 10.1107/S1744309109041086. Epub 2009 Oct 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19923743 Free PMC article.
-
Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.Structure. 1999 Aug 15;7(8):953-65. doi: 10.1016/s0969-2126(99)80122-1. Structure. 1999. PMID: 10467151
-
Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6.J Bacteriol. 2005 Aug;187(15):5067-74. doi: 10.1128/JB.187.15.5067-5074.2005. J Bacteriol. 2005. PMID: 16030198 Free PMC article.
-
Catechol dioxygenases.Essays Biochem. 1999;34:173-89. doi: 10.1042/bse0340173. Essays Biochem. 1999. PMID: 10730195 Review.
-
Mechanism of extradiol aromatic ring-cleaving dioxygenases.Curr Opin Struct Biol. 2008 Dec;18(6):644-9. doi: 10.1016/j.sbi.2008.11.001. Epub 2008 Nov 25. Curr Opin Struct Biol. 2008. PMID: 19007887 Free PMC article. Review.
Cited by
-
Proteins analysed as virtual knots.Sci Rep. 2017 Feb 13;7:42300. doi: 10.1038/srep42300. Sci Rep. 2017. PMID: 28205562 Free PMC article.
-
Iron acquisition system of Sphingobium sp. strain SYK-6, a degrader of lignin-derived aromatic compounds.Sci Rep. 2020 Jul 22;10(1):12177. doi: 10.1038/s41598-020-68984-2. Sci Rep. 2020. PMID: 32699224 Free PMC article.
-
Development and Application of Whole-Cell Biosensors for the Detection of Gallic Acid.ACS Synth Biol. 2023 Feb 17;12(2):533-543. doi: 10.1021/acssynbio.2c00537. Epub 2023 Feb 1. ACS Synth Biol. 2023. PMID: 36724292 Free PMC article.
-
Oxidative opening of the aromatic ring: Tracing the natural history of a large superfamily of dioxygenase domains and their relatives.J Biol Chem. 2019 Jun 28;294(26):10211-10235. doi: 10.1074/jbc.RA119.007595. Epub 2019 May 15. J Biol Chem. 2019. PMID: 31092555 Free PMC article.
-
Isolation of a novel platform bacterium for lignin valorization and its application in glucose-free cis,cis-muconate production.J Ind Microbiol Biotechnol. 2019 Aug;46(8):1071-1080. doi: 10.1007/s10295-019-02190-6. Epub 2019 May 27. J Ind Microbiol Biotechnol. 2019. PMID: 31134414
References
-
- Sterner R, Liebl W (2001) Thermophilic adaptation of proteins. Crit Rev Biochem Mol Biol 36: 39–106. - PubMed
-
- Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, et al. (1988) The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett 233: 129–133. - PubMed
-
- Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71: 1–15. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

