Multi-locus typing of Histomonas meleagridis isolates demonstrates the existence of two different genotypes
- PMID: 24658534
- PMCID: PMC3962415
- DOI: 10.1371/journal.pone.0092438
Multi-locus typing of Histomonas meleagridis isolates demonstrates the existence of two different genotypes
Abstract
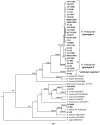
Histomonas meleagridis is the aetiological agent of histomonosis or "blackhead disease". Histomonosis is of special importance today, because there is no effective treatment to prevent its occurrence with considerable losses for the poultry industry. Despite its importance only a few molecular studies have yet been performed to investigate the degree of genetic diversity between different isolates of this parasite. In the present study a collection of well defined samples, previously shown positive for the DNA of H. meleagridis, was used to investigate genetic relatedness of the parasite. Samples originated from 25 turkey flocks collected in France between 2007 and 2010. Additionally, diagnostic samples, collected at our Clinic in Vienna, from different European countries and Azerbaijan, during 2010 to 2013 were included in the analyses. For the analysis three different genetic loci were analyzed: 18S rRNA, α-actinin1 and rpb1 genes. To amplify partial sequences of α-actinin1 and rpb1 genes, primers specifically targeting H. meleagridis were designed. Following PCR, the sequences of 18S rRNA, α-actinin1 and rpb1 loci were analyzed. Phylogenetic analyses demonstrated separation of H. meleagridis isolates in two different clusters. The majority of isolates grouped within the cluster 1 and originated from different European countries. The cluster 2 was rare and predominantly found in samples originating from France. Considering that the genetic variability of clusters can be seen as two distinct genetic types we propose the term genotype instead of cluster.
Conflict of interest statement
Figures




Similar articles
-
Prevalence and Genetic Characterization of Histomonas meleagridis in Chickens in Vietnam.Avian Dis. 2015 Jun;59(2):309-14. doi: 10.1637/10964-102414-Reg. Avian Dis. 2015. PMID: 26473683
-
Molecular biological features of strains of Histomonas meleagridis.Parasitol Res. 2009 Apr;104(5):1137-40. doi: 10.1007/s00436-008-1299-3. Epub 2008 Dec 11. Parasitol Res. 2009. PMID: 19082624
-
PCR and serology confirm the infection of turkey hens and their resilience to histomonosis in mixed flocks following high mortalities in toms.Parasit Vectors. 2019 May 14;12(1):228. doi: 10.1186/s13071-019-3482-z. Parasit Vectors. 2019. PMID: 31088526 Free PMC article.
-
Histomonas meleagridis--new insights into an old pathogen.Vet Parasitol. 2015 Feb 28;208(1-2):67-76. doi: 10.1016/j.vetpar.2014.12.018. Epub 2014 Dec 23. Vet Parasitol. 2015. PMID: 25576442 Review.
-
Unravelling the Immunity of Poultry Against the Extracellular Protozoan Parasite Histomonas meleagridis Is a Cornerstone for Vaccine Development: A Review.Front Immunol. 2018 Nov 2;9:2518. doi: 10.3389/fimmu.2018.02518. eCollection 2018. Front Immunol. 2018. PMID: 30450097 Free PMC article. Review.
Cited by
-
What is the importance of zoonotic trichomonads for human health?Trends Parasitol. 2014 Jul;30(7):333-41. doi: 10.1016/j.pt.2014.05.005. Epub 2014 Jun 18. Trends Parasitol. 2014. PMID: 24951156 Free PMC article. Review.
-
18S rRNA gene metabarcoding for investigation of gastrointestinal parasite diversity in great cormorants.Sci Rep. 2025 May 15;15(1):16954. doi: 10.1038/s41598-025-01774-w. Sci Rep. 2025. PMID: 40374688 Free PMC article.
-
Commensal or pathogen - a challenge to fulfil Koch's Postulates.Br Poult Sci. 2017 Feb;58(1):1-12. doi: 10.1080/00071668.2016.1245849. Br Poult Sci. 2017. PMID: 27724044 Free PMC article.
-
Histomonosis in Poultry: A Comprehensive Review.Front Vet Sci. 2022 May 6;9:880738. doi: 10.3389/fvets.2022.880738. eCollection 2022. Front Vet Sci. 2022. PMID: 35601402 Free PMC article. Review.
-
Host-specific targets of Histomonas meleagridis antigens revealed by immunoprecipitation.Sci Rep. 2025 Feb 17;15(1):5800. doi: 10.1038/s41598-025-88855-y. Sci Rep. 2025. PMID: 39962091 Free PMC article.
References
-
- Tyzzer EE (1920) The flagellate character and reclassification of the parasite producing "Blackhead" in turkeys- Histomonas (gen. nov.) meleagridis (Smith). J Parasitol 6: 124–131.
-
- McDougald LR (2005) Blackhead disease (histomoniasis) in poultry: a critical review. Avian Dis 49: 462–476. - PubMed
-
- Esquenet C, De Herdt P, De Bosschere H, Ronsmans S, Ducatelle R, et al. (2003) An outbreak of histomoniasis in free-range layer hens. Avian Pathol 32: 305–308. - PubMed
-
- van der Heijden HM, Landman WJ, Greve S, Peek R (2006) Genotyping of Histomonas meleagridis isolates based on Internal Transcribed Spacer-1 sequences. Avian Pathol 35: 330–334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

