Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy
- PMID: 24659329
- PMCID: PMC4001373
- DOI: 10.1105/tpc.113.121731
Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy
Abstract
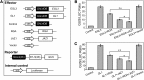
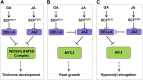
Integration of diverse environmental and endogenous signals to coordinately regulate growth, development, and defense is essential for plants to survive in their natural habitat. The hormonal signals gibberellin (GA) and jasmonate (JA) antagonistically and synergistically regulate diverse aspects of plant growth, development, and defense. GA and JA synergistically induce initiation of trichomes, which assist seed dispersal and act as barriers to protect plants against insect attack, pathogen infection, excessive water loss, and UV irradiation. However, the molecular mechanism underlying such synergism between GA and JA signaling remains unclear. In this study, we revealed a mechanism for GA and JA signaling synergy and identified a signaling complex of the GA pathway in regulation of trichome initiation. Molecular, biochemical, and genetic evidence showed that the WD-repeat/bHLH/MYB complex acts as a direct target of DELLAs in the GA pathway and that both DELLAs and JAZs interacted with the WD-repeat/bHLH/MYB complex to mediate synergism between GA and JA signaling in regulating trichome development. GA and JA induce degradation of DELLAs and JASMONATE ZIM-domain proteins to coordinately activate the WD-repeat/bHLH/MYB complex and synergistically and mutually dependently induce trichome initiation. This study provides deep insights into the molecular mechanisms for integration of different hormonal signals to synergistically regulate plant development.
Figures


References
-
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G.T., Genschik P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19: 1188–1193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

