Transcription of angiogenin and ribonuclease 4 is regulated by RNA polymerase III elements and a CCCTC binding factor (CTCF)-dependent intragenic chromatin loop
- PMID: 24659782
- PMCID: PMC4007445
- DOI: 10.1074/jbc.M114.551762
Transcription of angiogenin and ribonuclease 4 is regulated by RNA polymerase III elements and a CCCTC binding factor (CTCF)-dependent intragenic chromatin loop
Abstract
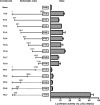
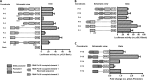
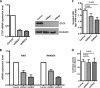
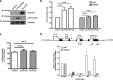
Angiogenin (ANG) and ribonuclease 4 (RNASE4), two members of the secreted and vertebrate-specific ribonuclease superfamily, play important roles in cancers and neurodegenerative diseases. The ANG and RNASE4 genes share genetic regions with promoter activities, but the structure and regulation of these putative promotes are unknown. We have characterized the promoter regions, defined the transcription start site, and identified a mechanism of transcription regulation that involves both RNA polymerase III (Pol III) elements and CCCTC binding factor (CTCF) sites. We found that two Pol III elements within the promoter region influence ANG and RNASE4 expression in a position- and orientation-dependent manner. We also provide evidence for the presence of an intragenic chromatin loop between the two CTCF binding sites located in two introns flanking the ANG coding exon. We found that formation of this intragenic loop preferentially enhances ANG transcription. These results suggest a multilayer transcriptional regulation of ANG and RNASE4 gene locus. These data also add more direct evidence to the notion that Pol III elements are able to directly influence Pol II gene transcription. Furthermore, our data indicate that a CTCF-dependent chromatin loop is able to differentially regulate transcription of genes that share the same promoters.
Keywords: Angiogenin; Chromatin; Promoters; RNA Polymerase; Ribonuclease; Rinonuclease 4; Transcription.
Figures







References
-
- Riordan J. F. (2001) Angiogenin. Methods Enzymol. 341, 263–273 - PubMed
-
- Subramanian V., Feng Y. (2007) A new role for angiogenin in neurite growth and pathfinding: implications for amyotrophic lateral sclerosis. Hum. Mol. Genet 16, 1445–1453 - PubMed
-
- Tello-Montoliu A., Patel J. V., Lip G. Y. (2006) Angiogenin: a review of the pathophysiology and potential clinical applications. J. Thromb. Haemost. 4, 1864–1874 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

