Dynamic regulation and function of histone monoubiquitination in plants
- PMID: 24659991
- PMCID: PMC3952079
- DOI: 10.3389/fpls.2014.00083
Dynamic regulation and function of histone monoubiquitination in plants
Abstract
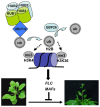
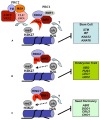
Polyubiquitin chain deposition on a target protein frequently leads to proteasome-mediated degradation whereas monoubiquitination modifies target protein property and function independent of proteolysis. Histone monoubiquitination occurs in chromatin and is in nowadays recognized as one critical type of epigenetic marks in eukaryotes. While H2A monoubiquitination (H2Aub1) is generally associated with transcription repression mediated by the Polycomb pathway, H2Bub1 is involved in transcription activation. H2Aub1 and H2Bub1 levels are dynamically regulated via deposition and removal by specific enzymes. We review knows and unknowns of dynamic regulation of H2Aub1 and H2Bub1 deposition and removal in plants and highlight the underlying crucial functions in gene transcription, cell proliferation/differentiation, and plant growth and development. We also discuss crosstalks existing between H2Aub1 or H2Bub1 and different histone methylations for an ample mechanistic understanding.
Keywords: Arabidopsis thaliana; chromatin; epigenetics; histone monoubiquitination; plant development; transcription regulation; ubiquitin.
Figures
References
-
- Berr A., McCallum E. J., Alioua A., Heintz D., Heitz T., Shen W.-H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP 8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154 1403–1414 10.1105/tpc.110.079962 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

