Developing a new production host from a blueprint: Bacillus pumilus as an industrial enzyme producer
- PMID: 24661794
- PMCID: PMC3987833
- DOI: 10.1186/1475-2859-13-46
Developing a new production host from a blueprint: Bacillus pumilus as an industrial enzyme producer
Abstract
Background: Since volatile and rising cost factors such as energy, raw materials and market competitiveness have a significant impact on the economic efficiency of biotechnological bulk productions, industrial processes need to be steadily improved and optimized. Thereby the current production hosts can undergo various limitations. To overcome those limitations and in addition increase the diversity of available production hosts for future applications, we suggest a Production Strain Blueprinting (PSB) strategy to develop new production systems in a reduced time lapse in contrast to a development from scratch.To demonstrate this approach, Bacillus pumilus has been developed as an alternative expression platform for the production of alkaline enzymes in reference to the established industrial production host Bacillus licheniformis.
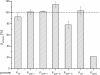
Results: To develop the selected B. pumilus as an alternative production host the suggested PSB strategy was applied proceeding in the following steps (dedicated product titers are scaled to the protease titer of Henkel's industrial production strain B. licheniformis at lab scale): Introduction of a protease production plasmid, adaptation of a protease production process (44%), process optimization (92%) and expression optimization (114%). To further evaluate the production capability of the developed B. pumilus platform, the target protease was substituted by an α-amylase. The expression performance was tested under the previously optimized protease process conditions and under subsequently adapted process conditions resulting in a maximum product titer of 65% in reference to B. licheniformis protease titer.
Conclusions: In this contribution the applied PSB strategy performed very well for the development of B. pumilus as an alternative production strain. Thereby the engineered B. pumilus expression platform even exceeded the protease titer of the industrial production host B. licheniformis by 14%. This result exhibits a remarkable potential of B. pumilus to be the basis for a next generation production host, since the strain has still a large potential for further genetic engineering. The final amylase titer of 65% in reference to B. licheniformis protease titer suggests that the developed B. pumilus expression platform is also suitable for an efficient production of non-proteolytic enzymes reaching a final titer of several grams per liter without complex process modifications.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

