Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia
- PMID: 24662245
- PMCID: PMC4016990
- DOI: 10.1038/ncomms4469
Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia
Abstract
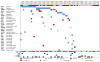
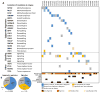
Relapsed paediatric acute lymphoblastic leukaemia (ALL) has high rates of treatment failure. Epigenetic regulators have been proposed as modulators of chemoresistance, here, we sequence genes encoding epigenetic regulators in matched diagnosis-remission-relapse ALL samples. We find significant enrichment of mutations in epigenetic regulators at relapse with recurrent somatic mutations in SETD2, CREBBP, MSH6, KDM6A and MLL2, mutations in signalling factors are not enriched. Somatic alterations in SETD2, including frameshift and nonsense mutations, are present at 12% in a large de novo ALL patient cohort. We conclude that the enrichment of mutations in epigenetic regulators at relapse is consistent with a role in mediating therapy resistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

