Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans
- PMID: 24662282
- PMCID: PMC3984390
- DOI: 10.1038/ncomms4483
Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans
Abstract
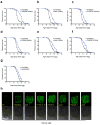
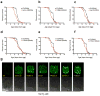
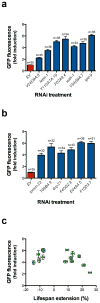
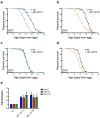
Recent studies have propagated the model that the mitochondrial unfolded protein response (UPR(mt)) is causal for lifespan extension from inhibition of the electron transport chain (ETC) in Caenorhabditis elegans. Here we report a genome-wide RNAi screen for negative regulators of the UPR(mt). Lifespan analysis of nineteen RNAi clones that induce the hsp-6p::gfp reporter demonstrate differential effects on longevity. Deletion of atfs-1, which is required for induction of the UPR(mt), fails to prevent lifespan extension from knockdown of two genes identified in our screen or following knockdown of the ETC gene cco-1. RNAi knockdown of atfs-1 also has no effect on lifespan extension caused by mutation of the ETC gene isp-1. Constitutive activation of the UPR(mt) by gain of function mutations in atfs-1 fails to extend lifespan. These observations identify several new factors that promote mitochondrial homoeostasis and demonstrate that the UPR(mt), as currently defined, is neither necessary nor sufficient for lifespan extension.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


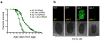
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

