Identifying mRNA sequence elements for target recognition by human Argonaute proteins
- PMID: 24663241
- PMCID: PMC4009607
- DOI: 10.1101/gr.162230.113
Identifying mRNA sequence elements for target recognition by human Argonaute proteins
Abstract
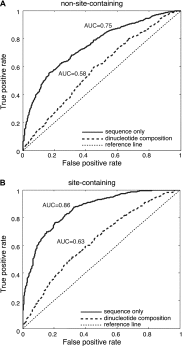
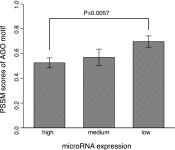
It is commonly known that mammalian microRNAs (miRNAs) guide the RNA-induced silencing complex (RISC) to target mRNAs through the seed-pairing rule. However, recent experiments that coimmunoprecipitate the Argonaute proteins (AGOs), the central catalytic component of RISC, have consistently revealed extensive AGO-associated mRNAs that lack seed complementarity with miRNAs. We herein test the hypothesis that AGO has its own binding preference within target mRNAs, independent of guide miRNAs. By systematically analyzing the data from in vivo cross-linking experiments with human AGOs, we have identified a structurally accessible and evolutionarily conserved region (∼10 nucleotides in length) that alone can accurately predict AGO-mRNA associations, independent of the presence of miRNA binding sites. Within this region, we further identified an enriched motif that was replicable on independent AGO-immunoprecipitation data sets. We used RNAcompete to enumerate the RNA-binding preference of human AGO2 to all possible 7-mer RNA sequences and validated the AGO motif in vitro. These findings reveal a novel function of AGOs as sequence-specific RNA-binding proteins, which may aid miRNAs in recognizing their targets with high specificity.
Figures

Similar articles
-
Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants.BMC Genomics. 2014 Dec 2;15(1):1049. doi: 10.1186/1471-2164-15-1049. BMC Genomics. 2014. PMID: 25443390 Free PMC article.
-
Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins.RNA. 2012 Nov;18(11):2041-55. doi: 10.1261/rna.035675.112. Epub 2012 Sep 27. RNA. 2012. PMID: 23019594 Free PMC article.
-
Target binding triggers hierarchical phosphorylation of human Argonaute-2 to promote target release.Elife. 2022 May 31;11:e76908. doi: 10.7554/eLife.76908. Elife. 2022. PMID: 35638597 Free PMC article.
-
Why Argonaute is needed to make microRNA target search fast and reliable.Semin Cell Dev Biol. 2017 May;65:20-28. doi: 10.1016/j.semcdb.2016.05.017. Epub 2016 May 26. Semin Cell Dev Biol. 2017. PMID: 27235676 Review.
-
MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions.Mol Cells. 2016 May 31;39(5):375-81. doi: 10.14348/molcells.2016.0013. Epub 2016 Apr 27. Mol Cells. 2016. PMID: 27117456 Free PMC article. Review.
Cited by
-
TarPmiR: a new approach for microRNA target site prediction.Bioinformatics. 2016 Sep 15;32(18):2768-75. doi: 10.1093/bioinformatics/btw318. Epub 2016 May 20. Bioinformatics. 2016. PMID: 27207945 Free PMC article.
-
A deep learning method for miRNA/isomiR target detection.Sci Rep. 2022 Jun 23;12(1):10618. doi: 10.1038/s41598-022-14890-8. Sci Rep. 2022. PMID: 35739186 Free PMC article.
-
tRForest: a novel random forest-based algorithm for tRNA-derived fragment target prediction.NAR Genom Bioinform. 2022 May 30;4(2):lqac037. doi: 10.1093/nargab/lqac037. eCollection 2022 Jun. NAR Genom Bioinform. 2022. PMID: 35664803 Free PMC article.
-
Roles of microRNA-140 in stem cell-associated early stage breast cancer.World J Stem Cells. 2014 Nov 26;6(5):591-7. doi: 10.4252/wjsc.v6.i5.591. World J Stem Cells. 2014. PMID: 25426255 Free PMC article. Review.
-
Expression Status of PIWIL1 as a Prognostic Marker of Colorectal Cancer.Dis Markers. 2017;2017:1204937. doi: 10.1155/2017/1204937. Epub 2017 May 29. Dis Markers. 2017. PMID: 28634417 Free PMC article.
References
-
- Bernhart SH, Hofacker IL, Stadler PF 2006. Local RNA base pairing probabilities in large sequences. Bioinformatics 22: 614–615 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
