Esterase LpEst1 from Lactobacillus plantarum: a novel and atypical member of the αβ hydrolase superfamily of enzymes
- PMID: 24663330
- PMCID: PMC3963902
- DOI: 10.1371/journal.pone.0092257
Esterase LpEst1 from Lactobacillus plantarum: a novel and atypical member of the αβ hydrolase superfamily of enzymes
Abstract
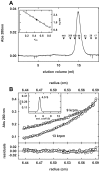
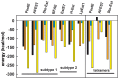
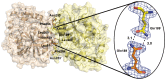
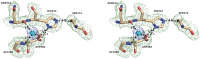
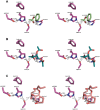
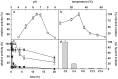
The genome of the lactic acid bacterium Lactobacillus plantarum WCFS1 reveals the presence of a rich repertoire of esterases and lipases highlighting their important role in cellular metabolism. Among them is the carboxylesterase LpEst1 a bacterial enzyme related to the mammalian hormone-sensitive lipase, which is known to play a central role in energy homeostasis. In this study, the crystal structure of LpEst1 has been determined at 2.05 Å resolution; it exhibits an αβ-hydrolase fold, consisting of a central β-sheet surrounded by α-helices, endowed with novel topological features. The structure reveals a dimeric assembly not comparable with any other enzyme from the bacterial hormone-sensitive lipase family, probably echoing the specific structural features of the participating subunits. Biophysical studies including analytical gel filtration and ultracentrifugation support the dimeric nature of LpEst1. Structural and mutational analyses of the substrate-binding pocket and active site together with biochemical studies provided insights for understanding the substrate profile of LpEst1 and suggested for the first time the conserved Asp173, which is adjacent to the nucleophile, as a key element in the stabilization of the loop where the oxyanion hole resides.
Conflict of interest statement
Figures



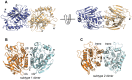
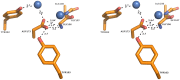
References
-
- Fojan P, Jonson PH, Petersen MTN, Petersen SB (2000) What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 82: 1033–1041. - PubMed
-
- Reis P, Holmberg K, Watke H, Leser ME, Miller R (2009) Lipases at interfaces: A review. Adv Colloid Interf Sci 147–148: 237–250. - PubMed
-
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, et al. (1991) Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253: 872–879. - PubMed
-
- Zhu X, Larsen NA, Basran A, Bruce NC, Wilson IA (2003) Observation of an arsenic adduct in an acetyl esterase crystal structure. J Biol Chem 278: 2008–2014. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

