Accumulation of N-acetylglucosamine oligomers in the plant cell wall affects plant architecture in a dose-dependent and conditional manner
- PMID: 24664205
- PMCID: PMC4012587
- DOI: 10.1104/pp.113.233742
Accumulation of N-acetylglucosamine oligomers in the plant cell wall affects plant architecture in a dose-dependent and conditional manner
Abstract
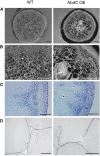
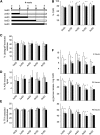
To study the effect of short N-acetylglucosamine (GlcNAc) oligosaccharides on the physiology of plants, N-ACETYLGLUCOSAMINYLTRANSFERASE (NodC) of Azorhizobium caulinodans was expressed in Arabidopsis (Arabidopsis thaliana). The corresponding enzyme catalyzes the polymerization of GlcNAc and, accordingly, β-1,4-GlcNAc oligomers accumulated in the plant. A phenotype characterized by difficulties in developing an inflorescence stem was visible when plants were grown for several weeks under short-day conditions before transfer to long-day conditions. In addition, a positive correlation between the oligomer concentration and the penetrance of the phenotype was demonstrated. Although NodC overexpression lines produced less cell wall compared with wild-type plants under nonpermissive conditions, no indications were found for changes in the amount of the major cell wall polymers. The effect on the cell wall was reflected at the transcriptome level. In addition to genes encoding cell wall-modifying enzymes, a whole set of genes encoding membrane-coupled receptor-like kinases were differentially expressed upon GlcNAc accumulation, many of which encoded proteins with an extracellular Domain of Unknown Function26. Although stress-related genes were also differentially expressed, the observed response differed from that of a classical chitin response. This is in line with the fact that the produced chitin oligomers were too small to activate the chitin receptor-mediated signal cascade. Based on our observations, we propose a model in which the oligosaccharides modify the architecture of the cell wall by acting as competitors in carbohydrate-carbohydrate or carbohydrate-protein interactions, thereby affecting noncovalent interactions in the cell wall or at the interface between the cell wall and the plasma membrane.
Figures





References
-
- Barker D, Bessou P, Jankowska E, Pages B, Stacey MJ. (1975) Distribution of static and dynamic gamma axons to cat intrafusal muscle-fibers. J Anat 119: 199–200
-
- Barny MA, Schoonejans E, Economou A, Johnston AWB, Downie JA. (1996) The C-terminal domain of the Rhizobium leguminosarum chitin synthase NodC is important for function and determines the orientation of the N-terminal region in the inner membrane. Mol Microbiol 19: 443–453 - PubMed
-
- Becker B, Holtgrefe S, Jung S, Wunrau C, Kandlbinder A, Baier M, Dietz KJ, Backhausen JE, Scheibe R. (2006) Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta 224: 380–393 - PubMed
-
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

