Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing
- PMID: 24664487
- PMCID: PMC3983811
- DOI: 10.1634/theoncologist.2013-0180
Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing
Abstract
Purpose: The success of precision oncology relies on accurate and sensitive molecular profiling. The Ion AmpliSeq Cancer Panel, a targeted enrichment method for next-generation sequencing (NGS) using the Ion Torrent platform, provides a fast, easy, and cost-effective sequencing workflow for detecting genomic "hotspot" regions that are frequently mutated in human cancer genes. Most recently, the U.K. has launched the AmpliSeq sequencing test in its National Health Service. This study aimed to evaluate the clinical application of the AmpliSeq methodology.
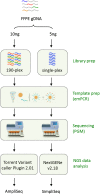
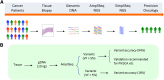
Methods: We used 10 ng of genomic DNA from formalin-fixed, paraffin-embedded human colorectal cancer (CRC) tumor specimens to sequence 46 cancer genes using the AmpliSeq platform. In a validation study, we developed an orthogonal NGS-based resequencing approach (SimpliSeq) to assess the AmpliSeq variant calls.
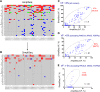
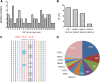
Results: Validated mutational analyses revealed that AmpliSeq was effective in profiling gene mutations, and that the method correctly pinpointed "true-positive" gene mutations with variant frequency >5% and demonstrated high-level molecular heterogeneity in CRC. However, AmpliSeq enrichment and NGS also produced several recurrent "false-positive" calls in clinically druggable oncogenes such as PIK3CA.
Conclusion: AmpliSeq provided highly sensitive and quantitative mutation detection for most of the genes on its cancer panel using limited DNA quantities from formalin-fixed, paraffin-embedded samples. For those genes with recurrent "false-positive" variant calls, caution should be used in data interpretation, and orthogonal verification of mutations is recommended for clinical decision making.
Keywords: AmpliSeq; Ion Torrent; Molecular diagnostics; Mutation; Next-generation sequencing; Precision oncology.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures
Comment in
-
In reply.Oncologist. 2015 May;20(5):e12. doi: 10.1634/theoncologist.2014-0357. Epub 2015 Apr 10. Oncologist. 2015. PMID: 25862746 Free PMC article. No abstract available.
-
Semiconductor-based sequencing of formalin-fixed, paraffin-embedded colorectal cancer samples.Oncologist. 2015 May;20(5):e10-1. doi: 10.1634/theoncologist.2014-0280. Epub 2015 Apr 10. Oncologist. 2015. PMID: 25862747 Free PMC article. No abstract available.
References
-
- Beadling C, Neff TL, Heinrich MC, et al. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mol Diagn. 2013;15:171–176. - PubMed
-
- Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. - PubMed
-
- Next-generation screening goes national in UK Cancer Discov. 2013;3:OF6.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

