Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation
- PMID: 24667637
- PMCID: PMC4001530
- DOI: 10.1172/JCI69592
Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation
Abstract
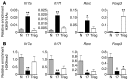
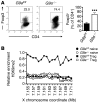
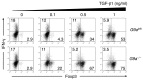
Inflammatory bowel disease (IBD) pathogenesis is associated with dysregulated CD4⁺ Th cell responses, with intestinal homeostasis depending on the balance between IL-17-producing Th17 and Foxp3⁺ Tregs. Differentiation of naive T cells into Th17 and Treg subsets is associated with specific gene expression profiles; however, the contribution of epigenetic mechanisms to controlling Th17 and Treg differentiation remains unclear. Using a murine T cell transfer model of colitis, we found that T cell-intrinsic expression of the histone lysine methyltransferase G9A was required for development of pathogenic T cells and intestinal inflammation. G9A-mediated dimethylation of histone H3 lysine 9 (H3K9me2) restricted Th17 and Treg differentiation in vitro and in vivo. H3K9me2 was found at high levels in naive Th cells and was lost following Th cell activation. Loss of G9A in naive T cells was associated with increased chromatin accessibility and heightened sensitivity to TGF-β1. Pharmacological inhibition of G9A methyltransferase activity in WT T cells promoted Th17 and Treg differentiation. Our data indicate that G9A-dependent H3K9me2 is a homeostatic epigenetic checkpoint that regulates Th17 and Treg responses by limiting chromatin accessibility and TGF-β1 responsiveness, suggesting G9A as a therapeutic target for treating intestinal inflammation.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

