FreeContact: fast and free software for protein contact prediction from residue co-evolution
- PMID: 24669753
- PMCID: PMC3987048
- DOI: 10.1186/1471-2105-15-85
FreeContact: fast and free software for protein contact prediction from residue co-evolution
Abstract
Background: 20 years of improved technology and growing sequences now renders residue-residue contact constraints in large protein families through correlated mutations accurate enough to drive de novo predictions of protein three-dimensional structure. The method EVfold broke new ground using mean-field Direct Coupling Analysis (EVfold-mfDCA); the method PSICOV applied a related concept by estimating a sparse inverse covariance matrix. Both methods (EVfold-mfDCA and PSICOV) are publicly available, but both require too much CPU time for interactive applications. On top, EVfold-mfDCA depends on proprietary software.
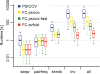
Results: Here, we present FreeContact, a fast, open source implementation of EVfold-mfDCA and PSICOV. On a test set of 140 proteins, FreeContact was almost eight times faster than PSICOV without decreasing prediction performance. The EVfold-mfDCA implementation of FreeContact was over 220 times faster than PSICOV with negligible performance decrease. EVfold-mfDCA was unavailable for testing due to its dependency on proprietary software. FreeContact is implemented as the free C++ library "libfreecontact", complete with command line tool "freecontact", as well as Perl and Python modules. All components are available as Debian packages. FreeContact supports the BioXSD format for interoperability.
Conclusions: FreeContact provides the opportunity to compute reliable contact predictions in any environment (desktop or cloud).
Figures
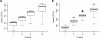
References
-
- Rost B, Sander C. Bridging the protein sequence-structure gap by structure predictions. Annual review of biophysics and biomolecular structure. 1996;25:113–136. - PubMed
-
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjölander K, Ferrin TE, Burley SK, Sali A. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39(Database issue):D465–474. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

