Should pharmacologists care about alternative splicing? IUPHAR Review 4
- PMID: 24670145
- PMCID: PMC3952800
- DOI: 10.1111/bph.12526
Should pharmacologists care about alternative splicing? IUPHAR Review 4
Abstract
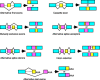
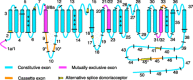
Alternative splicing of mRNAs occurs in the majority of human genes, and most differential splicing results in different protein isoforms with possibly different functional properties. However, there are many reported splicing variations that may be quite rare, and not all combinatorially possible variants of a given gene are expressed at significant levels. Genes of interest to pharmacologists are frequently expressed at such low levels that they are not adequately represented in genome-wide studies of transcription. In single-gene studies, data are commonly available on the relative abundance and functional significance of individual alternatively spliced exons, but there are rarely data that quantitate the relative abundance of full-length transcripts and define which combinations of exons are significant. A number of criteria for judging the significance of splice variants and suggestions for their nomenclature are discussed.
Keywords: 5-HT4 receptor; CaV1.2; CaV3.1; PAC1 receptor; alternative splicing; splice variant nomenclature.
Published 2013. This article is a U.S. Government work and is in the public domain in the USA.
Figures

References
-
- Bender E, Pindon A, van Oers I, Zhang YB, Gommeren W, Verhasselt P, et al. Structure of the human serotonin 5-HT4 receptor gene and cloning of a novel 5-HT4 splice variant. J Neurochem. 2000;74:478–489. - PubMed
-
- Brattelid T, Kvingedal AM, Krobert KA, Andressen KW, Bach T, Hystad ME, et al. Cloning, pharmacological characterisation and tissue distribution of a novel 5-HT4 receptor splice variant, 5-HT4(i) Naunyn Schmiedebergs Arch Pharmacol. 2004;369:616–628. - PubMed
-
- Cheng X, Pachuau J, Blaskova E, Asuncion-Chin M, Liu J, Dopico AM, et al. Alternative splicing of CaV1.2 channel exons in smooth muscle cells of resistance-size arteries generates currents with unique electrophysiological properties. Am J Physiol Heart Circ Physiol. 2009;297:H680–H688. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

