Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides
- PMID: 24670634
- PMCID: PMC4012432
- DOI: 10.1038/nature13110
Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides
Abstract
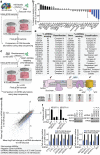
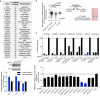
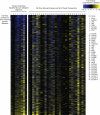
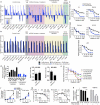
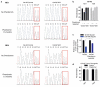
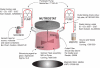
As the concentrations of highly consumed nutrients, particularly glucose, are generally lower in tumours than in normal tissues, cancer cells must adapt their metabolism to the tumour microenvironment. A better understanding of these adaptations might reveal cancer cell liabilities that can be exploited for therapeutic benefit. Here we developed a continuous-flow culture apparatus (Nutrostat) for maintaining proliferating cells in low-nutrient media for long periods of time, and used it to undertake competitive proliferation assays on a pooled collection of barcoded cancer cell lines cultured in low-glucose conditions. Sensitivity to low glucose varies amongst cell lines, and an RNA interference (RNAi) screen pinpointed mitochondrial oxidative phosphorylation (OXPHOS) as the major pathway required for optimal proliferation in low glucose. We found that cell lines most sensitive to low glucose are defective in the OXPHOS upregulation that is normally caused by glucose limitation as a result of either mitochondrial DNA (mtDNA) mutations in complex I genes or impaired glucose utilization. These defects predict sensitivity to biguanides, antidiabetic drugs that inhibit OXPHOS, when cancer cells are grown in low glucose or as tumour xenografts. Notably, the biguanide sensitivity of cancer cells with mtDNA mutations was reversed by ectopic expression of yeast NDI1, a ubiquinone oxidoreductase that allows bypass of complex I function. Thus, we conclude that mtDNA mutations and impaired glucose utilization are potential biomarkers for identifying tumours with increased sensitivity to OXPHOS inhibitors.
Figures




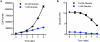

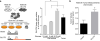

References
-
- Hirayama A, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. - PubMed
-
- Gullino PM, Grantham FH, Courtney AH. Glucose consumption by transplanted tumors in vivo. Cancer Res. 1967;27:1031–1040. - PubMed
-
- El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. - PubMed
-
- Seo BB, Matsuno-Yagi A, Yagi T. Modulation of oxidative phosphorylation of human kidney 293 cells by transfection with the internal rotenone-insensitive NADH-quinone oxidoreductase (NDI1) gene of Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1412:56–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

