An atlas of active enhancers across human cell types and tissues
- PMID: 24670763
- PMCID: PMC5215096
- DOI: 10.1038/nature12787
An atlas of active enhancers across human cell types and tissues
Abstract
Enhancers control the correct temporal and cell-type-specific activation of gene expression in multicellular eukaryotes. Knowing their properties, regulatory activity and targets is crucial to understand the regulation of differentiation and homeostasis. Here we use the FANTOM5 panel of samples, covering the majority of human tissues and cell types, to produce an atlas of active, in vivo-transcribed enhancers. We show that enhancers share properties with CpG-poor messenger RNA promoters but produce bidirectional, exosome-sensitive, relatively short unspliced RNAs, the generation of which is strongly related to enhancer activity. The atlas is used to compare regulatory programs between different cells at unprecedented depth, to identify disease-associated regulatory single nucleotide polymorphisms, and to classify cell-type-specific and ubiquitous enhancers. We further explore the utility of enhancer redundancy, which explains gene expression strength rather than expression patterns. The online FANTOM5 enhancer atlas represents a unique resource for studies on cell-type-specific enhancers and gene regulation.
Figures


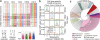
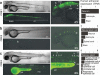

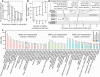
References
-
- Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–245. - PubMed
-
- Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Kodzius R, et al. CAGE: cap analysis of gene expression. Nature methods. 2006;3:211–222. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

