Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients
- PMID: 24671083
- PMCID: PMC4139721
- DOI: 10.1038/ismej.2014.28
Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients
Abstract
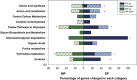
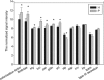
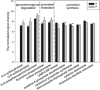
Determining the composition and function of subgingival dental plaque is crucial to understanding human periodontal health and disease, but it is challenging because of the complexity of the interactions between human microbiomes and human body. Here, we examined the phylogenetic and functional gene differences between periodontal and healthy individuals using MiSeq sequencing of 16S rRNA gene amplicons and a specific functional gene array (a combination of GeoChip 4.0 for biogeochemical processes and HuMiChip 1.0 for human microbiomes). Our analyses indicated that the phylogenetic and functional gene structure of the oral microbiomes were distinctly different between periodontal and healthy groups. Also, 16S rRNA gene sequencing analysis indicated that 39 genera were significantly different between healthy and periodontitis groups, and Fusobacterium, Porphyromonas, Treponema, Filifactor, Eubacterium, Tannerella, Hallella, Parvimonas, Peptostreptococcus and Catonella showed higher relative abundances in the periodontitis group. In addition, functional gene array data showed that a lower gene number but higher signal intensity of major genes existed in periodontitis, and a variety of genes involved in virulence factors, amino acid metabolism and glycosaminoglycan and pyrimidine degradation were enriched in periodontitis, suggesting their potential importance in periodontal pathogenesis. However, the genes involved in amino acid synthesis and pyrimidine synthesis exhibited a significantly lower relative abundance compared with healthy group. Overall, this study provides new insights into our understanding of phylogenetic and functional gene structure of subgingival microbial communities of periodontal patients and their importance in pathogenesis of periodontitis.
Figures




References
-
- Abe K. Butyric acid induces apoptosis in both human monocytes and lymphocytes equivalently. J Oral Sci. 2012;54:7–14. - PubMed
-
- Aimetti M, Cacciatore S, Graziano A, Tenori L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics. 2012;8:465–474.
-
- Alonso De La Peña V, Diz Dios P, Tojo Sierra R. Relationship between lactate dehydrogenase activity in saliva and oral health status. Arch Oral Biol. 2007;52:911–915. - PubMed
-
- Amano A. Bacterial adhesins to host components in periodontitis. Periodontol 2000. 2010;52:12–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

