Designing a high-throughput somatic mutation profiling panel specifically for gynaecological cancers
- PMID: 24671188
- PMCID: PMC3966900
- DOI: 10.1371/journal.pone.0093451
Designing a high-throughput somatic mutation profiling panel specifically for gynaecological cancers
Abstract
Somatic mutations play a major role in tumour initiation and progression. The mutation status of a tumour may predict prognosis and guide targeted therapies. The majority of techniques to study oncogenic mutations require high quality and quantity DNA or are analytically challenging. Mass-spectrometry based mutation analysis however is a relatively simple and high-throughput method suitable for formalin-fixed, paraffin-embedded (FFPE) tumour material. Targeted gene panels using this technique have been developed for several types of cancer. These current cancer hotspot panels are not focussed on the genes that are most relevant in gynaecological cancers. In this study, we report the design and validation of a novel, mass-spectrometry based panel specifically for gynaecological malignancies and present the frequencies of detected mutations. Using frequency data from the online Catalogue of Somatic Mutations in Cancer, we selected 171 somatic hotspot mutations in the 13 most important genes for gynaecological cancers, being BRAF, CDKN2A, CTNNB1, FBXW7, FGFR2, FGFR3, FOXL2, HRAS, KRAS, NRAS, PIK3CA, PPP2R1A and PTEN. A total of 546 tumours (205 cervical, 227 endometrial, 89 ovarian, and 25 vulvar carcinomas) were used to test and validate our panel, and to study the prevalence and spectrum of somatic mutations in these types of cancer. The results were validated by testing duplicate samples and by allele-specific qPCR. The panel presented here using mass-spectrometry shows to be reproducible and high-throughput, and is usefull in FFPE material of low quality and quantity. It provides new possibilities for studying large numbers of gynaecological tumour samples in daily practice, and could be useful in guided therapy selection.
Conflict of interest statement
Figures
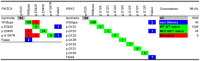
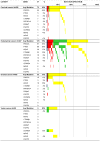
References
-
- Berek JS, Hacker NF (2010) Gynecologic Oncology. Philadelphia: Lippincott Williams & Wilkins.
-
- De RoockW, Claes B, Bernasconi D, De SchutterJ, Biesmans B, et al. (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 20 350: 2129–2139. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

